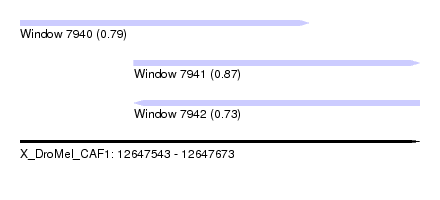
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,647,543 – 12,647,673 |
| Length | 130 |
| Max. P | 0.874274 |

| Location | 12,647,543 – 12,647,637 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 91.49 |
| Mean single sequence MFE | -24.54 |
| Consensus MFE | -24.24 |
| Energy contribution | -24.08 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.791993 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12647543 94 + 22224390 UUCAGAGUGAUUUCGGAAUUGCUUCCCUUCCAUUGGACGACAAAGGAGAGUUCCCAUCAAAGGACAUUCCUGGCC---GGCCCCUUUCAUUUGUCGU ....(((..(((....)))..))).((.......))((((((((.(((((...((.....(((.....)))....---))...))))).)))))))) ( -23.90) >DroSec_CAF1 1431 94 + 1 UCCAGAGUGAUUUCGGAAUUGCUUCCCUUCCAUCGGACGACAAAGGAGAGUCCCCAUCUUAGGACAUUCCUGGCC---GACCCCUUUCAUUUGUCGC (((.(((....)))((((.........))))...)))(((((((((((.((((........)))).)))))((..---....))......)))))). ( -23.70) >DroSim_CAF1 3290 94 + 1 UCCAGAGUGAUUUCGGAAUUGCUUCCCUUCCAUCGGACGACAAAGGAGAGUCCCCAUCAAAGGACAUUCCUGGCC---GGCCCCUUUCAUUUGUCGC (((.(((....)))((((.........))))...)))(((((((((((.((((........)))).)))))((..---..))........)))))). ( -24.80) >DroEre_CAF1 1471 94 + 1 UUCAGAGUGAUUUCCGAAUUGCUUCCUUCCCAUCGGACGACAAAGGAAAGUCCCCAUCAAAGGACAUUCCUGGCC---GGCCCCUUUCAUUUGUCCU ....(((..(((....)))..)))..........((((((...(((((.((((........)))).)))))((..---..))........)))))). ( -24.10) >DroYak_CAF1 3831 97 + 1 UCCAGAGUGAUUUCAGAAUUGCUUCCUUCCCAUCGAACGACAAAGGAAAGUCCCCAUCAAAGGACAUUCUUGGCCAUGGGCCCCUUUCAUUUGUCGU ....(((..(((....)))..)))............((((((((.((((((((........))))......((((...))))..)))).)))))))) ( -26.20) >consensus UCCAGAGUGAUUUCGGAAUUGCUUCCCUUCCAUCGGACGACAAAGGAGAGUCCCCAUCAAAGGACAUUCCUGGCC___GGCCCCUUUCAUUUGUCGU ....(((..(((....)))..))).............(((((((((((.((((........)))).)))))((((...))))........)))))). (-24.24 = -24.08 + -0.16)


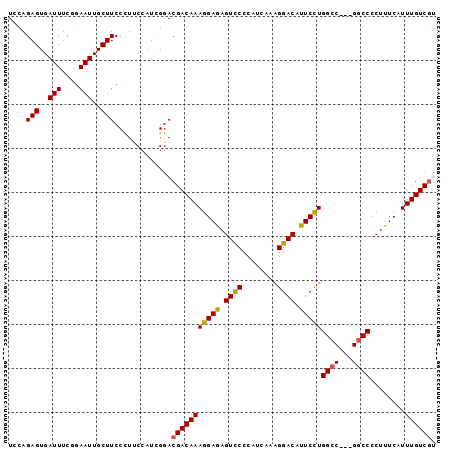
| Location | 12,647,580 – 12,647,673 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 87.16 |
| Mean single sequence MFE | -27.91 |
| Consensus MFE | -23.48 |
| Energy contribution | -23.36 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.874274 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12647580 93 + 22224390 CGACAAAGGAGAGUUCCCAUCAAAGGACAUUCCUGGCC---GGCCCCUUUCAUUUGUCGUAUUUCCAAUUUCGAGGGCCAGUCCAGGUCC---AUGGAC- (((((((.(((((...((.....(((.....)))....---))...))))).)))))))..........((((.(((((......)))))---.)))).- ( -27.50) >DroSec_CAF1 1468 96 + 1 CGACAAAGGAGAGUCCCCAUCUUAGGACAUUCCUGGCC---GACCCCUUUCAUUUGUCGCAUUUCCAAUUUCGAGGGCCAGUCCAGGUCCAUCAUGGAC- (((((((((((.((((........)))).)))))((..---....))......))))))....((((.....(((((((......))))).)).)))).- ( -27.90) >DroSim_CAF1 3327 96 + 1 CGACAAAGGAGAGUCCCCAUCAAAGGACAUUCCUGGCC---GGCCCCUUUCAUUUGUCGCAUUUCCAAUUUCGAGGGCCAGUCCAGGUCCAUCAUGGAC- (((((((((((.((((........)))).)))))((..---..))........))))))....((((.....(((((((......))))).)).)))).- ( -29.00) >DroEre_CAF1 1508 92 + 1 CGACAAAGGAAAGUCCCCAUCAAAGGACAUUCCUGGCC---GGCCCCUUUCAUUUGUCCUAUUUCCAAUUCCUAG-GCCAGUCCAAGUCC---GUGAGC- .(((..(((((.((((........)))).)))))(((.---((((........(((.........)))......)-))).)))...)))(---....).- ( -22.84) >DroYak_CAF1 3868 92 + 1 CGACAAAGGAAAGUCCCCAUCAAAGGACAUUCUUGGCCAUGGGCCCCUUUCAUUUGUCGUAUUUCCAAUUUGGAG-----GUCCAGGUAC---AUGGACC (((((((.((((((((........))))......((((...))))..)))).)))))))....(((.....)))(-----(((((.....---.)))))) ( -32.30) >consensus CGACAAAGGAGAGUCCCCAUCAAAGGACAUUCCUGGCC___GGCCCCUUUCAUUUGUCGUAUUUCCAAUUUCGAGGGCCAGUCCAGGUCC___AUGGAC_ (((((((((((.((((........)))).)))))((((...))))........)))))).....................(((((.........))))). (-23.48 = -23.36 + -0.12)

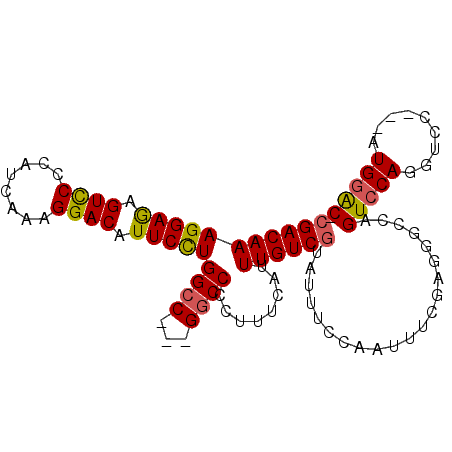
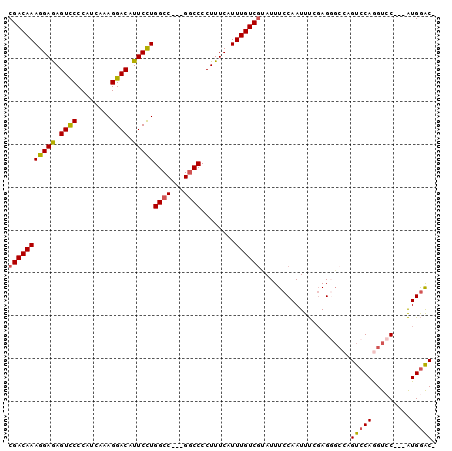
| Location | 12,647,580 – 12,647,673 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 87.16 |
| Mean single sequence MFE | -33.26 |
| Consensus MFE | -23.54 |
| Energy contribution | -24.02 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.734581 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12647580 93 - 22224390 -GUCCAU---GGACCUGGACUGGCCCUCGAAAUUGGAAAUACGACAAAUGAAAGGGGCC---GGCCAGGAAUGUCCUUUGAUGGGAACUCUCCUUUGUCG -......---((((((((.((((((((.....(((.........)))......))))))---)))))))....((((.....))))....)))....... ( -31.30) >DroSec_CAF1 1468 96 - 1 -GUCCAUGAUGGACCUGGACUGGCCCUCGAAAUUGGAAAUGCGACAAAUGAAAGGGGUC---GGCCAGGAAUGUCCUAAGAUGGGGACUCUCCUUUGUCG -......((..(((((((.((((((((.....(((.........)))......))))))---)))))))...(((((......))))).....))..)). ( -32.50) >DroSim_CAF1 3327 96 - 1 -GUCCAUGAUGGACCUGGACUGGCCCUCGAAAUUGGAAAUGCGACAAAUGAAAGGGGCC---GGCCAGGAAUGUCCUUUGAUGGGGACUCUCCUUUGUCG -......((..(((((((.((((((((.....(((.........)))......))))))---)))))))...((((((....)))))).....))..)). ( -35.70) >DroEre_CAF1 1508 92 - 1 -GCUCAC---GGACUUGGACUGGC-CUAGGAAUUGGAAAUAGGACAAAUGAAAGGGGCC---GGCCAGGAAUGUCCUUUGAUGGGGACUUUCCUUUGUCG -......---.(((.(((.(((((-((.....(((.........))).......)))))---)))))((((.((((((....)))))).))))...))). ( -31.60) >DroYak_CAF1 3868 92 - 1 GGUCCAU---GUACCUGGAC-----CUCCAAAUUGGAAAUACGACAAAUGAAAGGGGCCCAUGGCCAAGAAUGUCCUUUGAUGGGGACUUUCCUUUGUCG ((((((.---.....)))))-----)(((.....)))....(((((((.(((((.((((...)))).......(((((....)))))))))).))))))) ( -35.20) >consensus _GUCCAU___GGACCUGGACUGGCCCUCGAAAUUGGAAAUACGACAAAUGAAAGGGGCC___GGCCAGGAAUGUCCUUUGAUGGGGACUCUCCUUUGUCG .(((((.........)))))......................((((((.......((((...)))).(((..((((((....))))))..))))))))). (-23.54 = -24.02 + 0.48)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:34:48 2006