
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,633,556 – 12,633,657 |
| Length | 101 |
| Max. P | 0.557875 |

| Location | 12,633,556 – 12,633,657 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.24 |
| Mean single sequence MFE | -24.05 |
| Consensus MFE | -12.37 |
| Energy contribution | -13.22 |
| Covariance contribution | 0.85 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.557875 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12633556 101 + 22224390 G-CAAAGUUUUUGCGCCGAC-GAAAACCGGCAACUAUUAUGCGCUAAUUAUGACUAAAUAAUUGCAUGAAAUGCGC--------------CUAGCGCCAG-CC-AA-AACUCAACUCGAA .-...((((((.((((((..-......)))).........(((((((((((......))))))((((...))))..--------------..)))))..)-).-))-))))......... ( -25.20) >DroSec_CAF1 11866 82 + 1 G-CAAAGUUUUUGCGCCGAC-GAAAACCGGCAACUAUUAUGCGCUAAUUAUGACUAAAU---------------------------------AGCGCCAA-CC-AA-AACUCAACUCGAA (-(((.....))))((((..-......)))).........((((((.(((....))).)---------------------------------)))))...-..-..-............. ( -18.40) >DroSim_CAF1 14679 101 + 1 G-CAAAGUUUUUGCGCCGAC-GACAACCGGCAACUAUUAUGCGCUAAUUAUGACUAAAUAAUUGCAUGAAAUGCGC--------------CUAGCGCGAA-CC-AA-AACUCACCUCGAA (-(((.....))))((((..-......))))........((((((((((((......))))))((((...))))..--------------..))))))..-..-..-............. ( -22.70) >DroYak_CAF1 1974 101 + 1 G-CAAAGUU-UUGCGCCGAC-GAAAACCGGCAACUAUUAUGCGCUAAUUAUGACUAAAUAAUUGCAUGAAAUGCGC--------------CUAGCGCCAA-CC-AAAAACUCAACUCGAA (-(((....-))))((((..-......)))).........(((((((((((......))))))((((...))))..--------------..)))))...-..-................ ( -21.50) >DroAna_CAF1 4647 101 + 1 GGCAAAAGUUUUACGCCG----AAAAACGGCAACUAUUAUGCGCUAAUUAUGGCUAAAUAAUUGCAUGAAAUGCGC--------------CAAGCACCAAACCAAA-AACUCAACUCGAA (((...........((((----.....)))).....(((((((...(((((......)))))))))))).....))--------------)...............-............. ( -19.50) >DroPer_CAF1 6045 115 + 1 G-CAAAAGU-UGGCGUUGCCCAAAAAACGGCAACUAUUAUGCGCUAAUUAUGACUAAAUAAUUGCAUGAAAAGCGUCGGGCGCCGAGCGUCGAGCGCCAG-CC-AA-AGCUCAACUCGAA (-(.....(-((((((((((........))))))......(((((..(((((.(.........))))))..)))))..(((((((.....)).))))).)-))-))-.)).......... ( -37.00) >consensus G_CAAAGUUUUUGCGCCGAC_GAAAACCGGCAACUAUUAUGCGCUAAUUAUGACUAAAUAAUUGCAUGAAAUGCGC______________CUAGCGCCAA_CC_AA_AACUCAACUCGAA ..............((((.........)))).........(((((((((((......))))))(((.....)))..................)))))....................... (-12.37 = -13.22 + 0.85)

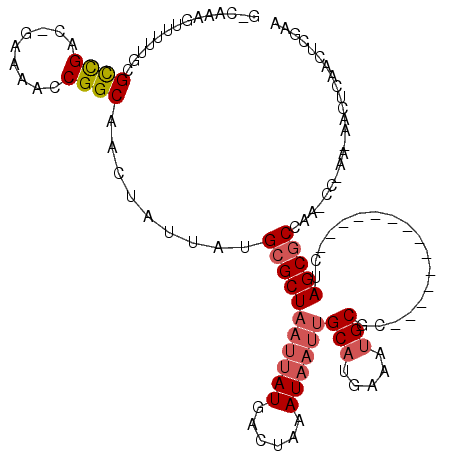
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:34:40 2006