| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,616,621 – 12,616,724 |
| Length | 103 |
| Max. P | 0.704294 |

| Location | 12,616,621 – 12,616,724 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 69.21 |
| Mean single sequence MFE | -18.20 |
| Consensus MFE | -8.19 |
| Energy contribution | -7.03 |
| Covariance contribution | -1.16 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.704294 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
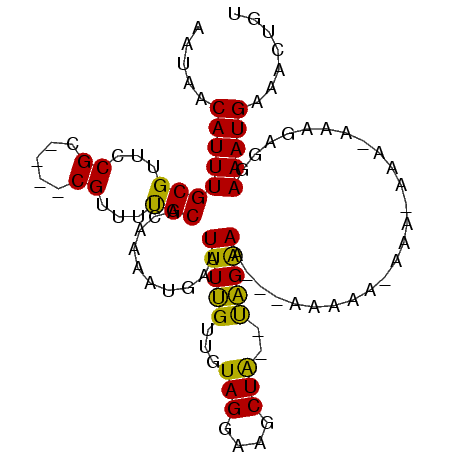
>X_DroMel_CAF1 12616621 103 - 22224390 AAUAACAUUUGCGUUCCGCUUUUCGUUUUGCACAAAAUGAAUUUUGUUGUAGGAAGCUACAUAGAAAUUAAAAAAAAAAUAAAGAAAGAGGAAAUGAAACUGU ....(((.((...((((.(((((((((((....))))))((((((..(((((....)))))..))))))..............))))).))))....)).))) ( -19.20) >DroSec_CAF1 5974 86 - 1 AAUAACAUUUGCGUU------------UCGCAGAAAAUGAAUUUUGUUGUAGGAAGCUA--UAGAAA---AAAAAGAAAAAAAAAAAGAGGAAAUGAAACUGU ....(((.((.((((------------((((((((......))))))(((((....)))--))....---....................)))))).)).))) ( -12.10) >DroEre_CAF1 6527 86 - 1 AAUAACAUUUGCGUUCCGCA--GCGUUUUGCGCAAAAUUAAUUUCGCUGUAGGUAACUG--CGGGAG---AAAAG----------CAAUGGAAAUGAAACUGU .....(((((.((((..((.--(((.....)))........((((.((((((....)))--))))))---)...)----------))))).)))))....... ( -23.30) >consensus AAUAACAUUUGCGUUCCGC____CGUUUUGCACAAAAUGAAUUUUGUUGUAGGAAGCUA__UAGAAA___AAAAA_AAA_AAA_AAAGAGGAAAUGAAACUGU .....((((((((...((.....))...)))..........(((((...(((....)))..))))).........................)))))....... ( -8.19 = -7.03 + -1.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:34:38 2006