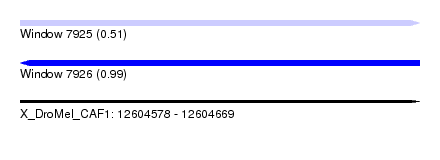
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,604,578 – 12,604,669 |
| Length | 91 |
| Max. P | 0.986657 |

| Location | 12,604,578 – 12,604,669 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 81.96 |
| Mean single sequence MFE | -11.10 |
| Consensus MFE | -9.23 |
| Energy contribution | -9.23 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.83 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.506399 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12604578 91 + 22224390 AAUCCAAACACACACACACACACAGCCAGCCACACACACUCAUAUCCAAAAAAUACGAAACACCUCAUGGCGUAGCUGAAGUGUCACGUUU .................((((.((((..((((.......((.(((.......))).)).........))))...))))..))))....... ( -11.89) >DroSec_CAF1 3321 73 + 1 ---CCAU------ACACACACGCAGCC------ACACACUCAUAUCC-AAAAAUACGAAACACCUCAUGGCGUAGCUGGAGUGCCACGU-- ---....------.....(((.((((.------......((.(((..-....))).)).((.((....)).)).))))..)))......-- ( -10.10) >DroSim_CAF1 4046 72 + 1 ---CCAU------ACACACACGCAGCC------ACACACUCAUAUC--AAAAAUACGAAACACCUCAUGGCGUAGCUGGAGUGUCACGU-- ---....------....((((.((((.------...........((--........)).((.((....)).)).))))..)))).....-- ( -11.30) >consensus ___CCAU______ACACACACGCAGCC______ACACACUCAUAUCC_AAAAAUACGAAACACCUCAUGGCGUAGCUGGAGUGUCACGU__ ..................(((.((((.............((.(((.......))).)).((.((....)).)).))))..)))........ ( -9.23 = -9.23 + -0.00)


| Location | 12,604,578 – 12,604,669 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 81.96 |
| Mean single sequence MFE | -26.00 |
| Consensus MFE | -21.44 |
| Energy contribution | -21.00 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.986657 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12604578 91 - 22224390 AAACGUGACACUUCAGCUACGCCAUGAGGUGUUUCGUAUUUUUUGGAUAUGAGUGUGUGUGGCUGGCUGUGUGUGUGUGUGUGUUUGGAUU ((((..(((((..((((((.((((((......((((((((.....))))))))....)))))))))))).....)))).)..))))..... ( -26.90) >DroSec_CAF1 3321 73 - 1 --ACGUGGCACUCCAGCUACGCCAUGAGGUGUUUCGUAUUUUU-GGAUAUGAGUGUGU------GGCUGCGUGUGUGU------AUGG--- --.((((((((.(((((((((((....)))..((((((((...-.))))))))...))------))))).).)))).)------))).--- ( -25.90) >DroSim_CAF1 4046 72 - 1 --ACGUGACACUCCAGCUACGCCAUGAGGUGUUUCGUAUUUUU--GAUAUGAGUGUGU------GGCUGCGUGUGUGU------AUGG--- --.((((((((.(((((((((((....)))..((((((((...--))))))))...))------))))).).)))).)------))).--- ( -25.20) >consensus __ACGUGACACUCCAGCUACGCCAUGAGGUGUUUCGUAUUUUU_GGAUAUGAGUGUGU______GGCUGCGUGUGUGU______AUGG___ .......((((.(((((((((((....)))))((((((((.....))))))))...........))))).).))))............... (-21.44 = -21.00 + -0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:34:33 2006