
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,599,133 – 12,599,270 |
| Length | 137 |
| Max. P | 0.994478 |

| Location | 12,599,133 – 12,599,236 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 79.49 |
| Mean single sequence MFE | -26.21 |
| Consensus MFE | -21.46 |
| Energy contribution | -21.80 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.48 |
| SVM RNA-class probability | 0.994478 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12599133 103 + 22224390 AUGAAAAAAAA--------------AAAUAGACAUCGAUGUUUCGGAAAAUCGAUAAAAACAUCGAUGCAUCGAUGUUAGCUUCGAUAUUUCGGUACCGCUAGUUGCCACGAAAGUC ..((((.....--------------.....(.(((((((((((((......)))....)))))))))))(((((........))))).))))((((.(....).))))((....)). ( -26.40) >DroSec_CAF1 7988 109 + 1 AUAAAUAUAAAUUGUCCGUAGUCUAUAAUAAACAUCGAUAUUUCGGGAAAUUGAUAAAAACAU--------CGAUGUUAACAUCGAUGUUUCGGCACCUCUAGUUGCCACGAAAGUC .....(((.((((..(((..(((.............)))....)))..)))).))).((((((--------(((((....))))))))))).((((.(....).))))((....)). ( -26.52) >DroSim_CAF1 7421 117 + 1 AUAAAUAUAAAUUGUCCGUAGUCUAUAAUAAACAUCGAUAUUUCGGGAAAUUGAUAAAAACAUCGCUGCAUCGAUGUUAUCAUCGAUGUUUCGGCACCUCUAGUUGCCACGAAAGUC .....((((.((((....)))).))))..((((((((((...((((....))))....(((((((......)))))))...)))))))))).((((.(....).))))((....)). ( -25.70) >consensus AUAAAUAUAAAUUGUCCGUAGUCUAUAAUAAACAUCGAUAUUUCGGGAAAUUGAUAAAAACAUCG_UGCAUCGAUGUUAACAUCGAUGUUUCGGCACCUCUAGUUGCCACGAAAGUC ....................................(((.(((((((((((((((...(((((((......)))))))...)))))).))))((((.(....).)))).)))))))) (-21.46 = -21.80 + 0.34)

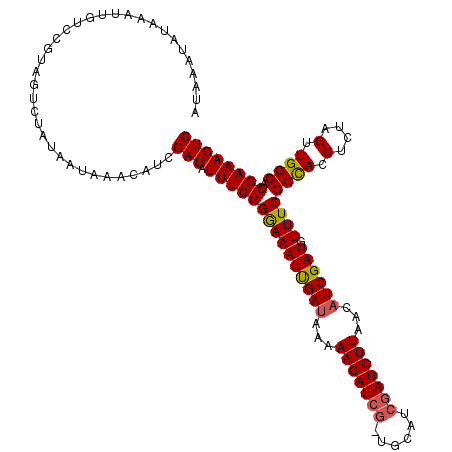

| Location | 12,599,133 – 12,599,236 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 79.49 |
| Mean single sequence MFE | -26.07 |
| Consensus MFE | -17.84 |
| Energy contribution | -19.07 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.881866 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12599133 103 - 22224390 GACUUUCGUGGCAACUAGCGGUACCGAAAUAUCGAAGCUAACAUCGAUGCAUCGAUGUUUUUAUCGAUUUUCCGAAACAUCGAUGUCUAUUU--------------UUUUUUUUCAU (((..(((((((.....))....(.((((.(((((....((((((((....))))))))....))))))))).)...)).))).))).....--------------........... ( -23.30) >DroSec_CAF1 7988 109 - 1 GACUUUCGUGGCAACUAGAGGUGCCGAAACAUCGAUGUUAACAUCG--------AUGUUUUUAUCAAUUUCCCGAAAUAUCGAUGUUUAUUAUAGACUACGGACAAUUUAUAUUUAU ...(((((.((((.(....).))))((((((((((((....)))))--------)))))))...........)))))..(((..(((((...)))))..)))............... ( -26.00) >DroSim_CAF1 7421 117 - 1 GACUUUCGUGGCAACUAGAGGUGCCGAAACAUCGAUGAUAACAUCGAUGCAGCGAUGUUUUUAUCAAUUUCCCGAAAUAUCGAUGUUUAUUAUAGACUACGGACAAUUUAUAUUUAU ....(((((((((.(....).)))).((((((((((((.(((((((......))))))).)))))........((....)))))))))..........))))).............. ( -28.90) >consensus GACUUUCGUGGCAACUAGAGGUGCCGAAACAUCGAUGAUAACAUCGAUGCA_CGAUGUUUUUAUCAAUUUCCCGAAAUAUCGAUGUUUAUUAUAGACUACGGACAAUUUAUAUUUAU ........(((((.(....).)))))(((((((((((..(((((((......)))))))....((........))..)))))))))))............................. (-17.84 = -19.07 + 1.23)

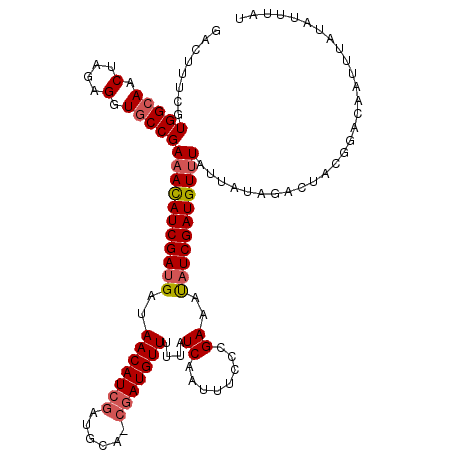
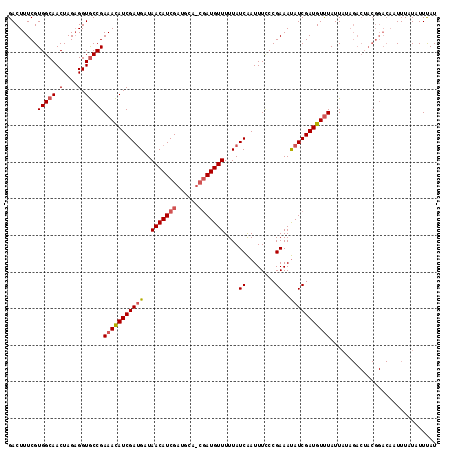
| Location | 12,599,159 – 12,599,270 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 89.79 |
| Mean single sequence MFE | -33.53 |
| Consensus MFE | -28.59 |
| Energy contribution | -28.93 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.912200 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12599159 111 + 22224390 UUUCGGAAAAUCGAUAAAAACAUCGAUGCAUCGAUGUUAGCUUCGAUAUUUCGGUACCGCUAGUUGCCACGAAAGUCGCUCGAACGCCGGUAACGGAAAUUCUCAGCAAAC ..((((((.(((((....((((((((....))))))))....))))).))))))....(((....((.((....)).))..(((..(((....)))...)))..))).... ( -33.80) >DroSec_CAF1 8028 103 + 1 UUUCGGGAAAUUGAUAAAAACAU--------CGAUGUUAACAUCGAUGUUUCGGCACCUCUAGUUGCCACGAAAGUCGCGCGAACGCCGGUAACGGAAAUUCUCAGCAAAC ....(((((..((....((((((--------(((((....)))))))))))...))..(((.((((((((....)).(((....))).)))))))))..)))))....... ( -32.70) >DroSim_CAF1 7461 111 + 1 UUUCGGGAAAUUGAUAAAAACAUCGCUGCAUCGAUGUUAUCAUCGAUGUUUCGGCACCUCUAGUUGCCACGAAAGUCGCGCGAACGCCGGUAACGGAAAUUCUCAGCAAAC ....(((((...............((((((((((((....)))))))))...)))...(((.((((((((....)).(((....))).)))))))))..)))))....... ( -34.10) >consensus UUUCGGGAAAUUGAUAAAAACAUCG_UGCAUCGAUGUUAACAUCGAUGUUUCGGCACCUCUAGUUGCCACGAAAGUCGCGCGAACGCCGGUAACGGAAAUUCUCAGCAAAC (((((((((((((((...(((((((......)))))))...)))))).))))((((.(....).)))).)))))...((..(((..(((....)))...)))...)).... (-28.59 = -28.93 + 0.34)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:34:28 2006