
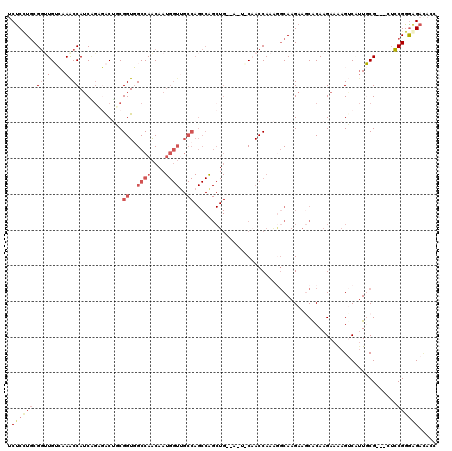
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,596,731 – 12,596,888 |
| Length | 157 |
| Max. P | 0.949660 |

| Location | 12,596,731 – 12,596,851 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.90 |
| Mean single sequence MFE | -37.65 |
| Consensus MFE | -11.02 |
| Energy contribution | -13.26 |
| Covariance contribution | 2.24 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.29 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.949660 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12596731 120 - 22224390 UCCCCUGCGGUUGUCAAACCAUCAGAGACUGCGGUGGCCAACAAUGGUUGCCAGCCAGCUGCAAGUGCAACCAAAGGCAAGAAGCACAAGAAAAGUCCUUGCGGGAGUCCGGGAGAAAAC (((((((.((((....))))..))).((((((((..((((....))))..)).))...(((((((..(........((.....)).........)..))))))).)))).))))...... ( -39.93) >DroSec_CAF1 5406 111 - 1 UCUCGUGCGGUUGUCAAGCCAUCGGAGACUUCGGUGGCCAACAAUGGUAGCCAGCCAGCUG------CAACCAAAGGCAAGAAGCACAAGAAAAGACAUUGCG---CUCCGGGAGACACC (((.((((..(((((..(((((((((...)))))))))......((((.((.((....)))------).))))..)))))...)))).)))............---.....(....)... ( -34.70) >DroSim_CAF1 4868 111 - 1 UCUCUUCCGGUUGUCAAGCCAUCGGAGACUUCGGUGGCCAACAAUGGUUGCCAGCCAGCUG------CAACCAAAGCCAAGAAGCACAAGAAAAGACAUUGCG---CUCCGGGAGACAUC ..((((((((((((...(((((((((...)))))))))..))))(((((((.((....)))------)))))).........(((.(((.........))).)---)))))))))).... ( -40.60) >DroEre_CAF1 6086 114 - 1 ACUCCUGCGGAUGGAAAACCAUCU---GCGCCGGUUGCCAACAAUGGUUGCCAGCCGGCUGUAAAUCCCACCAAGAGCAAGAAACGCAAGAAAAGUCCAUCCG---CUCCGGAGGAUACC .((((.(((((((((......(((---((((((((((.((((....)))).))))))))......((.......)))).)))..(....).....))))))))---)...))))...... ( -47.00) >DroYak_CAF1 5075 96 - 1 GCUCCUGCGCCUGGAAAACCAACAGCUACGGCGGUUGCCAGC---------------------AAUCCUACCAAGAGCAAGAAACACAAGAAGAGUCGUUCCG---CAGCGGGGGACACC .(((((((((.(((.....((((.((....)).)))))))))---------------------.............((..(((..((.......))..))).)---).)))))))..... ( -26.00) >consensus UCUCCUGCGGUUGUCAAACCAUCAGAGACUGCGGUGGCCAACAAUGGUUGCCAGCCAGCUG__A_U_CAACCAAAGGCAAGAAGCACAAGAAAAGUCAUUGCG___CUCCGGGAGACACC .((((((.((((....))))............((..((((....))))..)).........................................................))))))..... (-11.02 = -13.26 + 2.24)


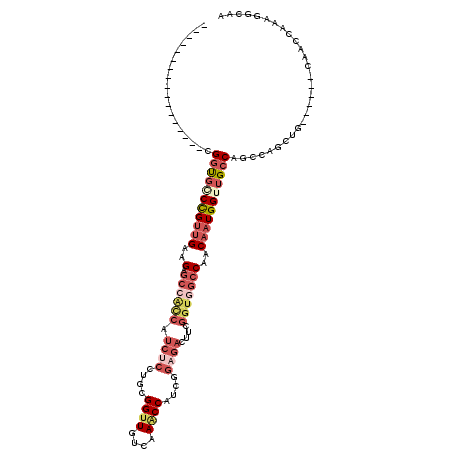
| Location | 12,596,771 – 12,596,888 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 68.29 |
| Mean single sequence MFE | -40.62 |
| Consensus MFE | -16.79 |
| Energy contribution | -20.73 |
| Covariance contribution | 3.94 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.41 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.751140 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12596771 117 - 22224390 UGCCGCUCCACCCAAUGCGAUGGCCGGUGAAGAGCA---CUCCCCUGCGGUUGUCAAACCAUCAGAGACUGCGGUGGCCAACAAUGGUUGCCAGCCAGCUGCAAGUGCAACCAAAGGCAA ((((.((.((((....((....)).)))).)).(((---((....((((((((.(.......(((...))).((..((((....))))..)).).))))))))))))).......)))). ( -41.40) >DroSec_CAF1 5443 114 - 1 UGCCGCGCCUCGCAACGCGGUGCCCGUUGAAGAGCCACCAUCUCGUGCGGUUGUCAAGCCAUCGGAGACUUCGGUGGCCAACAAUGGUAGCCAGCCAGCUG------CAACCAAAGGCAA .((((((........))))))((((((((..(.((((((.........((((....))))...(....)...)))))))..)))))(((((......))))------).......))).. ( -44.60) >DroSim_CAF1 4905 97 - 1 -----------------CGGUGCCCGUUGAAGAGCCACCAUCUCUUCCGGUUGUCAAGCCAUCGGAGACUUCGGUGGCCAACAAUGGUUGCCAGCCAGCUG------CAACCAAAGCCAA -----------------.(((....((((....((((((.(((((...((((....))))...)))))....))))))))))..(((((((.((....)))------))))))..))).. ( -37.90) >DroEre_CAF1 6123 97 - 1 -----------------CGGCGACUGUCGAAGA---GUCUACUCCUGCGGAUGGAAAACCAUCU---GCGCCGGUUGCCAACAAUGGUUGCCAGCCGGCUGUAAAUCCCACCAAGAGCAA -----------------..((((((.......)---)))(((....((((((((....))))))---))((((((((.((((....)))).)))))))).))).............)).. ( -38.60) >consensus _________________CGGUGCCCGUUGAAGAGCCACCAUCUCCUGCGGUUGUCAAACCAUCGGAGACUUCGGUGGCCAACAAUGGUUGCCAGCCAGCUG______CAACCAAAGGCAA ..................(((((((((((..(.((((((.((((....((((....))))....))))....)))))))..)))))))))))............................ (-16.79 = -20.73 + 3.94)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:34:25 2006