| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,408,974 – 1,409,083 |
| Length | 109 |
| Max. P | 0.982828 |

| Location | 1,408,974 – 1,409,083 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 67.00 |
| Mean single sequence MFE | -42.38 |
| Consensus MFE | -17.87 |
| Energy contribution | -17.73 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.42 |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.982828 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
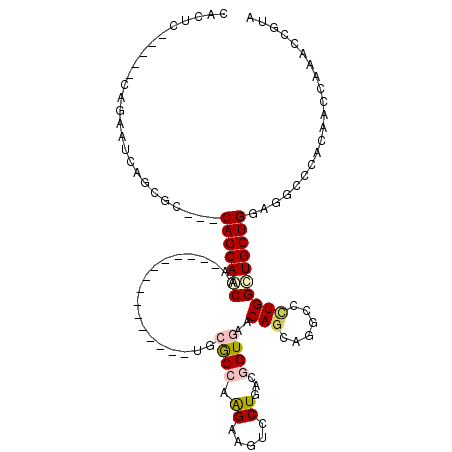
>X_DroMel_CAF1 1408974 109 + 22224390 GACGGUUUGGUUGCGCGCCGCCAGCAACCAGGGCCUGCUGUUCACCGACAGCACUUCCUGGCGCACCGCGACGGGCACAGCUGUUGCUGGUGGCGCUGAUUUUG-----GAGUG ..((.((..(...((((((((((((((((((((..(((((((....)))))))..)))))(.((.(((...))))).)....)))))))))))))).)...)..-----)).)) ( -54.00) >DroVir_CAF1 849 94 + 1 UACACUUUGAUUCUGGGCUUGCAGCAGCCACGGCUUUUUGUUCACCGUGAGGAUCUCUUGGCGAG---------------UGGCUGCUG---CAGCUGCUGUUGAGUGU--GUG (((((.(..((....((((.(((((((((((.(((....(....((....))....)..)))..)---------------)))))))))---)))))...))..)))))--).. ( -42.60) >DroPse_CAF1 45317 91 + 1 UACGGUUUGGUUCUGGGCCUCCAGCAGCCAAGGCAUGCUGUUUACUCUCAGGACUUCUUUGUGUC---------------UUGUUGCUG---UGAUUAAUUCUG-----AUUCA ((((((..(((((((((....(((((((....)).)))))......)))))))))..........---------------.....))))---))..........-----..... ( -23.53) >DroSec_CAF1 14443 109 + 1 GACGGUUUGGUUGCGCGCCGCCAGCAACCAGGGCCUGCUGUUCACCGACAGCACUUCCUGGCGCACCGCGACGGGCACAGUUGUUGCUGGUGGCGCUGAUUCUG-----GAGUG ..((.((..(...((((((((((((((((((((..(((((((....)))))))..)))))(.((.(((...))))).)....)))))))))))))).)...)..-----)).)) ( -55.30) >DroEre_CAF1 10775 109 + 1 GACGGUUUGGUUGCGCGCCGCCAGCAACCAGGGCCUGCUGUUCACCGACAGCACUUCCUGGCGCACCGCGACGGGCACAGCUGUUGCUGGUGGCGCUGAUUCUG-----GAGUG ..((.((..(...((((((((((((((((((((..(((((((....)))))))..)))))(.((.(((...))))).)....)))))))))))))).)...)..-----)).)) ( -55.30) >DroPer_CAF1 48097 91 + 1 UACGGUUUGGUUCUGGGCCUCCAGCAGCCAAGGCAUGCUGUUUACUCUCAGGACUUCUUUGUGUC---------------UUGUUGCUG---UGAUUAAUUCUG-----AUUCA ((((((..(((((((((....(((((((....)).)))))......)))))))))..........---------------.....))))---))..........-----..... ( -23.53) >consensus GACGGUUUGGUUCCGCGCCGCCAGCAACCAGGGCCUGCUGUUCACCGACAGCACUUCCUGGCGCA_______________UUGUUGCUG___GCGCUGAUUCUG_____GAGUG ...(((..(....)..)))..((((((((((((..(((((........)))))..))))).....((.....))........)))))))......................... (-17.87 = -17.73 + -0.14)

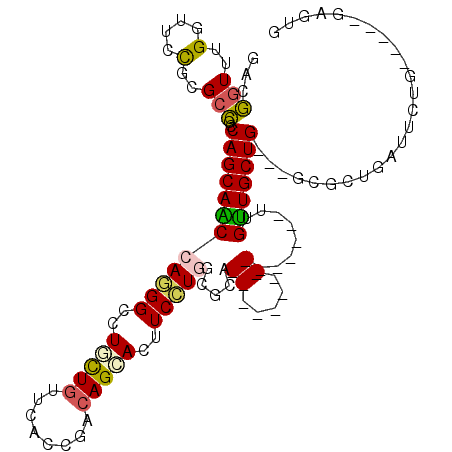
| Location | 1,408,974 – 1,409,083 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 67.00 |
| Mean single sequence MFE | -36.03 |
| Consensus MFE | -11.56 |
| Energy contribution | -11.92 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.32 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.849307 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1408974 109 - 22224390 CACUC-----CAAAAUCAGCGCCACCAGCAACAGCUGUGCCCGUCGCGGUGCGCCAGGAAGUGCUGUCGGUGAACAGCAGGCCCUGGUUGCUGGCGGCGCGCAACCAAACCGUC .....-----........(((((.((((((((....(((((((...))).))))((((...((((((......))))))...)))))))))))).))))).............. ( -48.70) >DroVir_CAF1 849 94 - 1 CAC--ACACUCAACAGCAGCUG---CAGCAGCCA---------------CUCGCCAAGAGAUCCUCACGGUGAACAAAAAGCCGUGGCUGCUGCAAGCCCAGAAUCAAAGUGUA ...--(((((........((((---(((((((((---------------((((((..(((...)))..)))))..........))))))))))).)))..........))))). ( -35.67) >DroPse_CAF1 45317 91 - 1 UGAAU-----CAGAAUUAAUCA---CAGCAACAA---------------GACACAAAGAAGUCCUGAGAGUAAACAGCAUGCCUUGGCUGCUGGAGGCCCAGAACCAAACCGUA .....-----............---..((.....---------------(((........)))(((.(.((...(((((.((....)))))))...)))))).........)). ( -17.20) >DroSec_CAF1 14443 109 - 1 CACUC-----CAGAAUCAGCGCCACCAGCAACAACUGUGCCCGUCGCGGUGCGCCAGGAAGUGCUGUCGGUGAACAGCAGGCCCUGGUUGCUGGCGGCGCGCAACCAAACCGUC .....-----........(((((.((((((((....(((((((...))).))))((((...((((((......))))))...)))))))))))).))))).............. ( -48.70) >DroEre_CAF1 10775 109 - 1 CACUC-----CAGAAUCAGCGCCACCAGCAACAGCUGUGCCCGUCGCGGUGCGCCAGGAAGUGCUGUCGGUGAACAGCAGGCCCUGGUUGCUGGCGGCGCGCAACCAAACCGUC .....-----........(((((.((((((((....(((((((...))).))))((((...((((((......))))))...)))))))))))).))))).............. ( -48.70) >DroPer_CAF1 48097 91 - 1 UGAAU-----CAGAAUUAAUCA---CAGCAACAA---------------GACACAAAGAAGUCCUGAGAGUAAACAGCAUGCCUUGGCUGCUGGAGGCCCAGAACCAAACCGUA .....-----............---..((.....---------------(((........)))(((.(.((...(((((.((....)))))))...)))))).........)). ( -17.20) >consensus CACUC_____CAGAAUCAGCGC___CAGCAACAA_______________UGCGCCAAGAAGUCCUGACGGUGAACAGCAGGCCCUGGCUGCUGGAGGCCCACAACCAAACCGUA .........................(((((((...................((((.((.....))...))))..(((......))))))))))..................... (-11.56 = -11.92 + 0.36)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:45:43 2006