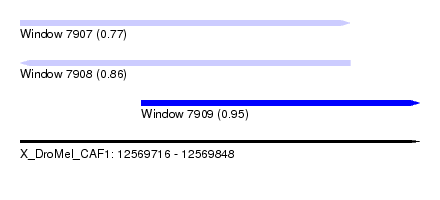
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,569,716 – 12,569,848 |
| Length | 132 |
| Max. P | 0.949021 |

| Location | 12,569,716 – 12,569,825 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 76.11 |
| Mean single sequence MFE | -25.17 |
| Consensus MFE | -15.96 |
| Energy contribution | -15.93 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.773810 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
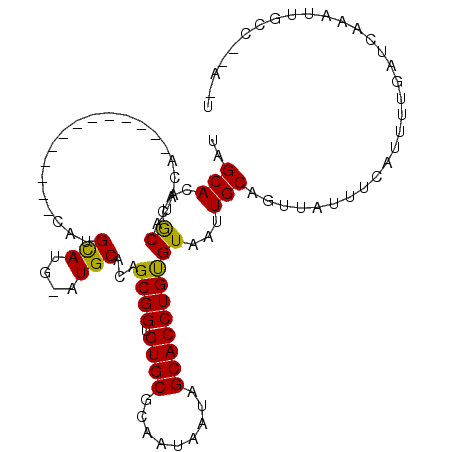
>X_DroMel_CAF1 12569716 109 + 22224390 AUUGGGGCAAUUUGAUCAAAAUGAAAUAACGGCAAUUACGCAGGUGCUAUUAUUGCGCACACCGCUGUGCAUGCAUACAUGUGUAGCGUCUAUAUGUAUGCGUGUGCUA ...((.((.((((.((....)).))))....(((((...((....))....)))))))...))...(..((((((((((((((.......))))))))))))))..).. ( -33.00) >DroEre_CAF1 37113 86 + 1 ----------UUUGAUCAAAAUGAAAUUACUGCAAUUACACAGGUGCUAUUAUUGCGCACACCGCUGUGCAUACAUGCAUG-------------UGUAUGCGUGUGCUA ----------.....................(((((..((....)).....)))))((((((......((((((((....)-------------))))))))))))).. ( -21.30) >DroYak_CAF1 25802 92 + 1 AAUAAGGCAAUUUGAUCAAAAUAAAAUAACAGCAAUUACACAGGUGCUAUUAUUGCGCACACCGCUGUGCA----UGCGUA-------------UGUAUGUGUGUGCUA .....(((.((((.....))))...............(((((.(((((((...((((((......))))))----...)))-------------.)))).)))))))). ( -21.20) >consensus A_U__GGCAAUUUGAUCAAAAUGAAAUAACAGCAAUUACACAGGUGCUAUUAUUGCGCACACCGCUGUGCAU_CAUGCAUG_____________UGUAUGCGUGUGCUA ...............................(((((..((....)).....)))))((((((.((.((((((.....................)))))))))))))).. (-15.96 = -15.93 + -0.02)


| Location | 12,569,716 – 12,569,825 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 76.11 |
| Mean single sequence MFE | -22.47 |
| Consensus MFE | -15.29 |
| Energy contribution | -14.63 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858498 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12569716 109 - 22224390 UAGCACACGCAUACAUAUAGACGCUACACAUGUAUGCAUGCACAGCGGUGUGCGCAAUAAUAGCACCUGCGUAAUUGCCGUUAUUUCAUUUUGAUCAAAUUGCCCCAAU ..(((...((((((((.(((...)))...)))))))).)))...((((.((((.........))))))))((((((...((((........))))..))))))...... ( -26.80) >DroEre_CAF1 37113 86 - 1 UAGCACACGCAUACA-------------CAUGCAUGUAUGCACAGCGGUGUGCGCAAUAAUAGCACCUGUGUAAUUGCAGUAAUUUCAUUUUGAUCAAA---------- ..(((((((((((((-------------......)))))))......))))))(((((.((((...))))...))))).....................---------- ( -21.70) >DroYak_CAF1 25802 92 - 1 UAGCACACACAUACA-------------UACGCA----UGCACAGCGGUGUGCGCAAUAAUAGCACCUGUGUAAUUGCUGUUAUUUUAUUUUGAUCAAAUUGCCUUAUU ..((.(((((.....-------------..(((.----......)))))))).))((((((((((..........))))))))))........................ ( -18.91) >consensus UAGCACACGCAUACA_____________CAUGCAUG_AUGCACAGCGGUGUGCGCAAUAAUAGCACCUGUGUAAUUGCAGUUAUUUCAUUUUGAUCAAAUUGCC__A_U ..(((...((.....................(((....)))...((((.((((.........))))))))))...)))............................... (-15.29 = -14.63 + -0.66)


| Location | 12,569,756 – 12,569,848 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 81.37 |
| Mean single sequence MFE | -29.03 |
| Consensus MFE | -20.60 |
| Energy contribution | -21.49 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.949021 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
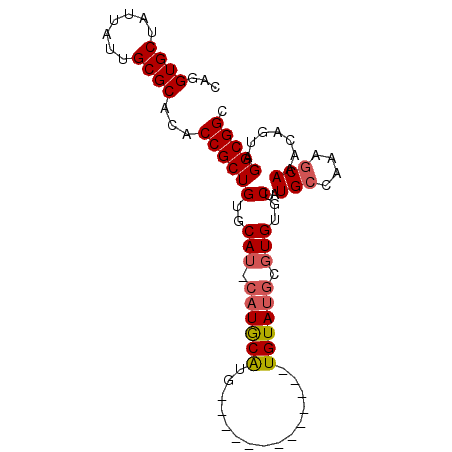
>X_DroMel_CAF1 12569756 92 + 22224390 CAGGUGCUAUUAUUGCGCACACCGCUGUGCAUGCAUACAUGUGUAGCGUCUAUAUGUAUGCGUGUGCUAUGCCAAUACAAACAGUAGGCGGC ...((((.......))))...((((((..((((((((((((((.......))))))))))))))..)........(((.....)))))))). ( -35.20) >DroEre_CAF1 37143 78 + 1 CAGGUGCUAUUAUUGCGCACACCGCUGUGCAUACAUGCAUG-------------UGUAUGCGUGUGCUAUGCCAAAGCAAACAGUAGGCGG- ...((((.......))))...(((((..((((((((....)-------------)))))))((.((((.......)))).))....)))))- ( -27.80) >DroYak_CAF1 25842 75 + 1 CAGGUGCUAUUAUUGCGCACACCGCUGUGCA----UGCGUA-------------UGUAUGUGUGUGCUAUGCCAAAGCAAACAGUAGGCGGC ...((((.......))))...((((((..((----(((((.-------------...)))))))..)..(((....))).......))))). ( -24.10) >consensus CAGGUGCUAUUAUUGCGCACACCGCUGUGCAU_CAUGCAUG_____________UGUAUGCGUGUGCUAUGCCAAAGCAAACAGUAGGCGGC ...((((.......))))...((((((..(((.((((((...............)))))).)))..)..(((....))).......))))). (-20.60 = -21.49 + 0.89)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:34:18 2006