
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,545,887 – 12,546,083 |
| Length | 196 |
| Max. P | 0.986820 |

| Location | 12,545,887 – 12,546,005 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.80 |
| Mean single sequence MFE | -53.50 |
| Consensus MFE | -39.43 |
| Energy contribution | -39.22 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.74 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.518777 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12545887 118 + 22224390 GCAGCCGCUUUGGCCACCUUUUCACCGGCAGCCGUUGCUUCGACAGCCUUAGUGGCGGCGCUUGCCUUUUCGGCGGCACUUUCAGCCGCUUUG-GCAGCAUCAGCUGCCGCUUUCUCU-G (((((.(((...(((...........)))))).))))).....(((....((((((((((((.(((.....((((((.......))))))..)-)))))....)))))))))....))-) ( -48.20) >DroPse_CAF1 55268 118 + 1 GCGGCUGCCUUGGCCACCUUCUCGCUGACGACAAUGGCCUCGACCGCCUUGGUGACGGCGCUGGCCUUCUCAGCGGCACUUUCGGCCGCCUUG-GCGGCAUCGGCGGCAGCCUUCUCG-G ..(((((((...(((......(((((((.((....((((.....((((..(....)))))..)))).)))))))))........(((((....-)))))...))))))))))......-. ( -56.00) >DroPer_CAF1 54577 120 + 1 GCGGCUGCCUUGGCCACCUUCUCGCUGACGACAAUGGCCUCGACCGCCUUGGUGACGGCGCUGGCCUUCUCAGCGGCACUUUCGGCCGCCUUGGGCGGCAUCGGCGGCAGCCUUCUCGGG ..(((((((...(((......(((((((.((....((((.....((((..(....)))))..)))).)))))))))........((((((...))))))...))))))))))........ ( -56.30) >consensus GCGGCUGCCUUGGCCACCUUCUCGCUGACGACAAUGGCCUCGACCGCCUUGGUGACGGCGCUGGCCUUCUCAGCGGCACUUUCGGCCGCCUUG_GCGGCAUCGGCGGCAGCCUUCUCG_G ((.((((((..((((......(((((((.((....((((.....((((........))))..)))).))))))))).......))))(((......)))...)))))).))......... (-39.43 = -39.22 + -0.21)



| Location | 12,545,887 – 12,546,005 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.80 |
| Mean single sequence MFE | -59.80 |
| Consensus MFE | -47.91 |
| Energy contribution | -47.03 |
| Covariance contribution | -0.87 |
| Combinations/Pair | 1.25 |
| Mean z-score | -3.27 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.986820 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12545887 118 - 22224390 C-AGAGAAAGCGGCAGCUGAUGCUGC-CAAAGCGGCUGAAAGUGCCGCCGAAAAGGCAAGCGCCGCCACUAAGGCUGUCGAAGCAACGGCUGCCGGUGAAAAGGUGGCCAAAGCGGCUGC .-.........((((((....)))))-)...(((((((.....((((((.....(((....)))..((((..(((.((((......)))).)))))))....)))))).....))))))) ( -53.90) >DroPse_CAF1 55268 118 - 1 C-CGAGAAGGCUGCCGCCGAUGCCGC-CAAGGCGGCCGAAAGUGCCGCUGAGAAGGCCAGCGCCGUCACCAAGGCGGUCGAGGCCAUUGUCGUCAGCGAGAAGGUGGCCAAGGCAGCCGC .-......(((((((..((..(((((-(..(((((((....).))))))..((.(((....))).)).....)))))))).(((((((.(((....)))...)))))))..))))))).. ( -63.20) >DroPer_CAF1 54577 120 - 1 CCCGAGAAGGCUGCCGCCGAUGCCGCCCAAGGCGGCCGAAAGUGCCGCUGAGAAGGCCAGCGCCGUCACCAAGGCGGUCGAGGCCAUUGUCGUCAGCGAGAAGGUGGCCAAGGCAGCCGC ........(((((((..((..((((((...(((((((....).))))))..((.(((....))).)).....)))))))).(((((((.(((....)))...)))))))..))))))).. ( -62.30) >consensus C_CGAGAAGGCUGCCGCCGAUGCCGC_CAAGGCGGCCGAAAGUGCCGCUGAGAAGGCCAGCGCCGUCACCAAGGCGGUCGAGGCCAUUGUCGUCAGCGAGAAGGUGGCCAAGGCAGCCGC ........((((((((((...(((......(((((((....).)))))).....)))...(((((((.....))))).)).))).......((((.(.....).))))...))))))).. (-47.91 = -47.03 + -0.87)


| Location | 12,545,927 – 12,546,044 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 82.62 |
| Mean single sequence MFE | -57.23 |
| Consensus MFE | -40.93 |
| Energy contribution | -41.83 |
| Covariance contribution | 0.90 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.829438 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
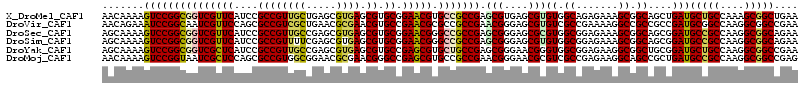
>X_DroMel_CAF1 12545927 117 - 22224390 GUGAGCGUGCGGAACGUGCCGCCGAGCGUGAGCGUGUGGCAGAGAAAGCGGCAGCUGAUGCUGCCAAAGCGGCUGAAAGUGCCGCCGAAAAGGCAAGCGCCGCCACUAAGGCUGUCG (((.(((.(((.....((((((((.((....))...)))).........((((((....))))))...(((((.......)))))......))))..)))))))))........... ( -52.70) >DroYak_CAF1 1156 117 - 1 GUGAGCGUGCCGAGCGUGCUGCCGAGCGGGAACGGGUGGCGGAGAAGGCGGCUGCGGAUGCUGCCAAGGCGGCCGAAAGUGCCGCCGAGAAGGCAAGCGCCGCUAGCAAGGCUGUCG ..((.((.(((..((.(.((((((..((....))..))))))...).)).(((((((.(((((((..(((((((....).)))))).....))).)))))))).)))..))))))). ( -60.60) >DroMoj_CAF1 1054 117 - 1 GCGAACGGGCCGAGCGUGCCGCCGAACGGGAACGCGUCGCCGAGAAGGCAGCCGCUGAUGCCGCCAAGGCGGCCGAGUGUGCCGCCGAGAAGGCGAGCGCCGUUAGCAAAGCUGCCG .......(((.((((((.(((.....)))..)))).))))).....((((((.(((((((((((....)))))...(.((((((((.....)))).)))))))))))...)))))). ( -58.40) >consensus GUGAGCGUGCCGAGCGUGCCGCCGAGCGGGAACGCGUGGCAGAGAAGGCGGCAGCUGAUGCUGCCAAGGCGGCCGAAAGUGCCGCCGAGAAGGCAAGCGCCGCUAGCAAGGCUGUCG ((.((((.((...((.((((((((..((....))..))))......((((((.......))))))..(((((((....).)))))).....)))).)))))))).)).......... (-40.93 = -41.83 + 0.90)


| Location | 12,545,967 – 12,546,083 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 83.22 |
| Mean single sequence MFE | -55.10 |
| Consensus MFE | -37.35 |
| Energy contribution | -37.30 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.577054 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12545967 116 - 22224390 AACAAAAGUCCGGCGGUCGUUCAUCCGCCGUUGCUGAGCGUGAGCGUGCGGAACGUGCCGCCGAGCGUGAGCGUGUGGCAGAGAAAGCGGCAGCUGAUGCUGCCAAAGCGGCUGAA ......((((((((((........))))))..(((..((....))..((......((((((...((....))..))))))......))((((((....))))))..)))))))... ( -52.30) >DroVir_CAF1 1247 116 - 1 AACAGAAAUCCGGCAAUCGUUCCAGCGCCGUCGCUGAACGCGAACGUGCCGAACGCGCCGCCGAACGGGAGCGUGUCGCCGAAAAGGCCGCCGCCGAUGCGGCCAAGGCGGCCGAA ..........(((((.((((..(((((....)))))...))))...)))))..((.((((((...(((((.....)).)))....((((((.......))))))..)))))))).. ( -51.80) >DroSec_CAF1 9843 116 - 1 AGCAAAAGUCCGGCGGUCGUUCAUCCGCCGUUGCCGAGCGUGAGCGUGCGGAACGGGCCGCCGAGCGGGAGCGCGUGGCGGAGAAAGCGGCAGCGGAUGCCGCCAAGGCGGCAGAA .......(((((((((........)))))((((((..((....))..((........((((((.(((....))).)))))).....))))))))))))(((((....))))).... ( -61.82) >DroSim_CAF1 8101 116 - 1 AGCAAAAGUCCGGCGGUCGUUCAUCCGCCGUUUUCGAGCGUGAGCGUGCGGAACGGGCCGCCGAGCGGGAGCGUGUGGCGGAGAAAGCGGCAGCGGAUGCCGCCAAGGCGGCAGAA .......(((((.(.((((((..(((.((((((.((.((....))...))))))))(((((...((....))..))))))))...)))))).))))))(((((....))))).... ( -57.50) >DroYak_CAF1 1196 116 - 1 AGCAAAAGUCCGGCGGUCGCUCAUCCGCCGUUGCCGAGCGUGAGCGUGCCGAGCGUGCUGCCGAGCGGGAACGGGUGGCGGAGAAGGCGGCUGCGGAUGCUGCCAAGGCGGCCGAA .......(((((.((((((((..(((((((((.(((..((..(((((((...)))))))..))..))).)))....))))))...)))))))))))))(((((....))))).... ( -59.40) >DroMoj_CAF1 1094 116 - 1 AACAAAAGUCCGGUAAUCGCUCCAGCGCCGUGGCGGAACGCGAACGGGCCGAGCGUGCCGCCGAACGGGAACGCGUCGCCGAGAAGGCAGCCGCUGAUGCCGCCAAGGCGGCCGAG ..........((((....((..((((((((((((((.((((...........)))).)))))..))))....((...(((.....))).)))))))..))(((....))))))).. ( -47.80) >consensus AACAAAAGUCCGGCGGUCGUUCAUCCGCCGUUGCCGAGCGUGAGCGUGCCGAACGUGCCGCCGAGCGGGAGCGUGUGGCGGAGAAAGCGGCAGCGGAUGCCGCCAAGGCGGCCGAA .......(((((((((((((((....((((((((.....)))).)).)).))))).))))))).(((....)))((.((.......)).))....)))(((((....))))).... (-37.35 = -37.30 + -0.05)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:34:07 2006