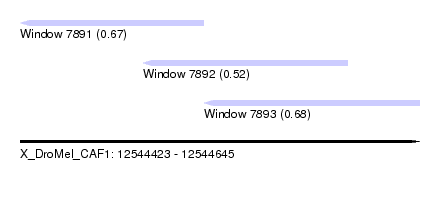
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,544,423 – 12,544,645 |
| Length | 222 |
| Max. P | 0.682747 |

| Location | 12,544,423 – 12,544,525 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 83.25 |
| Mean single sequence MFE | -35.28 |
| Consensus MFE | -22.49 |
| Energy contribution | -22.05 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.669769 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12544423 102 - 22224390 AGGAUGGAUCGAUGAUGGCCAAAACAUCGAUGGCAUCCACAUCGUCGGCAUCGGUUGUUGUCACAUCUACGCCAUCGGCAUCCUCAUCGUCAUUGUCAUCCG .((((((..(((((((((........((((((((.........(.(((((.....))))).)........))))))))........))))))))))))))). ( -32.52) >DroSec_CAF1 8367 93 - 1 AGGAUGGAUCGA---UGGCCAAGACAUCGAUGGCAUGCACGUCGUCGGCUUCGGUUGUCGUCACAUCCACGCCAUCGGCAGCCUCAUCGU------CAUCCG .((((((..(((---(((....((..((((((((......))))))))..))((((((((...............)))))))))))))))------))))). ( -36.26) >DroSim_CAF1 6644 93 - 1 AGGAUGGAUCGA---UGGCCAAGACAUCGAUGGCAUCCACGUCGUCGGCUUCGGUUGUCGUCACAUCCACGCCAUCGGCAGCCUCAUCGU------CAUCCG .((((((..(((---(((....((..((((((((......))))))))..))((((((((...............)))))))))))))))------))))). ( -36.26) >DroEre_CAF1 9623 93 - 1 AGGAUGCAUCGA---UGGCCAAGGCUUCGAUGCCGUCCACGUCGUCGACAUCCGCUGCCGUCGCAUACACGCCACCGGCCUCCCCGGCGU------CAUCCG .(((((...(((---((((...((..((((((.((....)).))))))...))...))))))).....(((((...((....)).)))))------))))). ( -36.10) >consensus AGGAUGGAUCGA___UGGCCAAGACAUCGAUGGCAUCCACGUCGUCGGCAUCGGUUGUCGUCACAUCCACGCCAUCGGCAGCCUCAUCGU______CAUCCG .(((((.........((((...(((.((((((((......)))))))).....)))...)))).......(((...))).................))))). (-22.49 = -22.05 + -0.44)

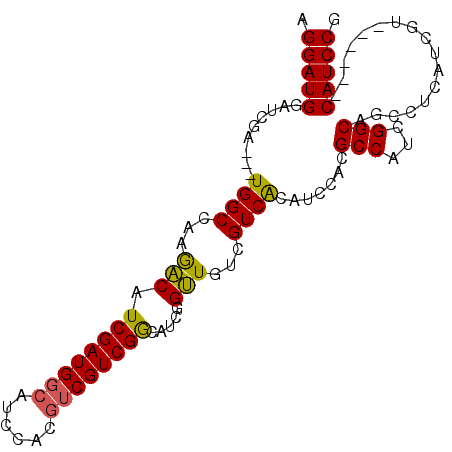
| Location | 12,544,491 – 12,544,605 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 90.52 |
| Mean single sequence MFE | -38.22 |
| Consensus MFE | -27.39 |
| Energy contribution | -27.95 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.72 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.523133 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12544491 114 - 22224390 UGCAACGUCAGCUAUGACCAAUAGCAGCACCAGUGGCAGCAUGAACAUCAGCAAUAGUCACGGAUUGAAGGCCAAUGCCAAGGAUGGAUCGAUGAUGGCCAAAACAUCGAUGGC .((..((((.(((((.....)))))....((.(((((.((.((.....))))....)))))))......(((....)))...)))).(((((((.(......).))))))).)) ( -29.40) >DroSec_CAF1 8429 111 - 1 CGCAACGGCGGCUAUGACCAAUAGCAGCACCAGUGGCAGCAUGAACAUCAGCAAUAGUCACGGCCUGAAGGCCAAUGCCAAGGAUGGAUCGA---UGGCCAAGACAUCGAUGGC (((....)))(((((.....))))).((.(((.(((((((.((.....)))).........((((....))))..)))))....)))(((((---((.......))))))).)) ( -39.30) >DroSim_CAF1 6706 111 - 1 CGCAACGGCGGCUAUGACCAAUAGCAGCACCAGUGGCAGCAUGAACAUCAGCAAUAGUCACGGCCUGAAGGCCAAUGCCAAGGAUGGAUCGA---UGGCCAAGACAUCGAUGGC (((....)))(((((.....))))).((.(((.(((((((.((.....)))).........((((....))))..)))))....)))(((((---((.......))))))).)) ( -39.30) >DroEre_CAF1 9685 111 - 1 CGCGGCGGCAGCUAUGGCCAAUAGCAGCGCCAGUGGCAGCAUGAGCAUCAGCAACAGUCACGGCCAGAAGGCCAGUGCCAAGGAUGCAUCGA---UGGCCAAGGCUUCGAUGCC ...((((.(.(((((.....))))).)))))..(((((((.((.....)))).........((((....))))..))))).....(((((((---.(((....)))))))))). ( -44.90) >consensus CGCAACGGCAGCUAUGACCAAUAGCAGCACCAGUGGCAGCAUGAACAUCAGCAAUAGUCACGGCCUGAAGGCCAAUGCCAAGGAUGGAUCGA___UGGCCAAGACAUCGAUGGC ......(((.(((((.....)))))....(((.(((((((.((.....)))).........((((....))))..)))))....)))..........))).............. (-27.39 = -27.95 + 0.56)

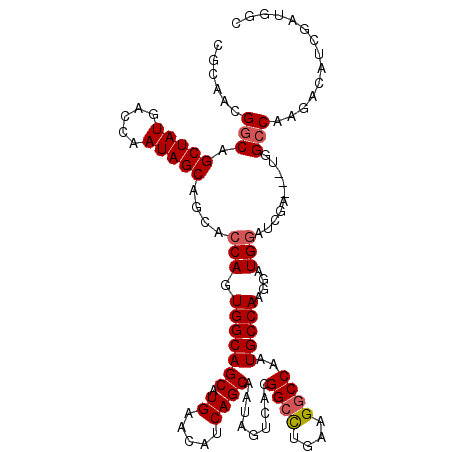

| Location | 12,544,525 – 12,544,645 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.28 |
| Mean single sequence MFE | -42.08 |
| Consensus MFE | -31.40 |
| Energy contribution | -32.77 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.682747 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12544525 120 - 22224390 GGAAACGAUUCGAACUGAAUUCCGGUGAGGCAGGUGGAAAUGCAACGUCAGCUAUGACCAAUAGCAGCACCAGUGGCAGCAUGAACAUCAGCAAUAGUCACGGAUUGAAGGCCAAUGCCA .....(((((((.((((.....))))...((.((((...((((.......(((((.....))))).((.......)).))))...)))).))........)))))))..(((....))). ( -32.30) >DroSec_CAF1 8460 120 - 1 GGAAGCGCUUCGAACUGAAUUCCGGUGAGGCAGCUGGGAACGCAACGGCGGCUAUGACCAAUAGCAGCACCAGUGGCAGCAUGAACAUCAGCAAUAGUCACGGCCUGAAGGCCAAUGCCA ......(((((..((((.....))))))))).(((((...(((....)))(((((.....)))))....)))))((((((.((.....)))).........((((....))))..)))). ( -42.10) >DroSim_CAF1 6737 120 - 1 GGAAACGCUUCGAACUGAAUUCCGGUGAGGCGGCUGGAAACGCAACGGCGGCUAUGACCAAUAGCAGCACCAGUGGCAGCAUGAACAUCAGCAAUAGUCACGGCCUGAAGGCCAAUGCCA .....((((((..((((.....))))))))))(((((...(((....)))(((((.....)))))....)))))((((((.((.....)))).........((((....))))..)))). ( -44.20) >DroEre_CAF1 9716 120 - 1 GGAAACGCUUCGAGCUGAAUGCGGCUGAGGCAGCAGGAAACGCGGCGGCAGCUAUGGCCAAUAGCAGCGCCAGUGGCAGCAUGAGCAUCAGCAACAGUCACGGCCAGAAGGCCAGUGCCA (....).......(((((.(((.((((..((.((.(....)))((((.(.(((((.....))))).))))).))..))))....))))))))....(.(((((((....)))).))).). ( -49.70) >consensus GGAAACGCUUCGAACUGAAUUCCGGUGAGGCAGCUGGAAACGCAACGGCAGCUAUGACCAAUAGCAGCACCAGUGGCAGCAUGAACAUCAGCAAUAGUCACGGCCUGAAGGCCAAUGCCA (....)(((((..((((.....))))))))).(((((...(((....)))(((((.....)))))....)))))((((((.((.....)))).........((((....))))..)))). (-31.40 = -32.77 + 1.38)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:34:04 2006