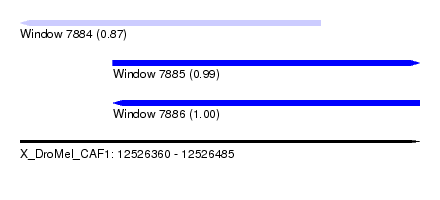
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,526,360 – 12,526,485 |
| Length | 125 |
| Max. P | 0.999024 |

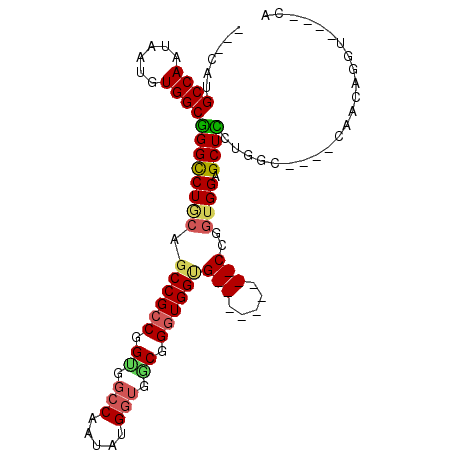
| Location | 12,526,360 – 12,526,454 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 73.47 |
| Mean single sequence MFE | -42.50 |
| Consensus MFE | -24.50 |
| Energy contribution | -25.25 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.871845 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12526360 94 - 22224390 ---CAUGCCAAUAAUGUGGCUGGUCUGCAGCCGCCGGUGGCCAAUAUGGGGACGGGUGGUGGUGUUCAACCAGGUGGAGCUGCUGGG----CAACAGGU----CA ---...((((......)))).(..(((((((((((....((((...((....))..))))(((.....))).))))..)))))..(.----...)))..----). ( -34.80) >DroSec_CAF1 55973 94 - 1 ---CACGCCAAUAAUGUGGCGGGCCUGCAGCCGCCGGUGGCCAAUAUGGUGCCGGGUGGUGGUGUCCAACCCGGUGGUGCUCCAGGC----CCACAGGU----CA ---...((((......))))(((((((.(((.(((....(((.....)))(((((((((......)).))))))))))))).)))))----))......----.. ( -47.80) >DroEre_CAF1 56088 85 - 1 ---CAUGCCAAUAAUGUGGCGGGCCUCCAGCCGCCGGCGGCCAAUAUGGUGGCGGCUGGAG---------CCGCUGGAGCUCCGGGC----CAGCAGCU----CA ---...((((......))))((((((((((((((((.((.......)).))))))))))))---------..(((((..(....).)----)))).)))----). ( -46.40) >DroYak_CAF1 56827 96 - 1 GGUCAUGCCAAUAAUGUGGCCGGCCUACAGCCGCCGGUGGGCAAUAUGGUGGCUGGUGGCG---------CUGGUGGUGCUGGUGGUGUCCAAGCGGGUGGAACA .((..((((....(..(.((((((((((.(((((((((.(.(.....).).))))))))).---------...)))).)))))).)..).......))))..)). ( -41.00) >consensus ___CAUGCCAAUAAUGUGGCGGGCCUGCAGCCGCCGGUGGCCAAUAUGGUGGCGGGUGGUG_________CCGGUGGAGCUCCUGGC____CAACAGGU____CA ......((((......))))((((((((.((((((.((.(((.....))).)).))))))(((.....)))..)))).))))....................... (-24.50 = -25.25 + 0.75)


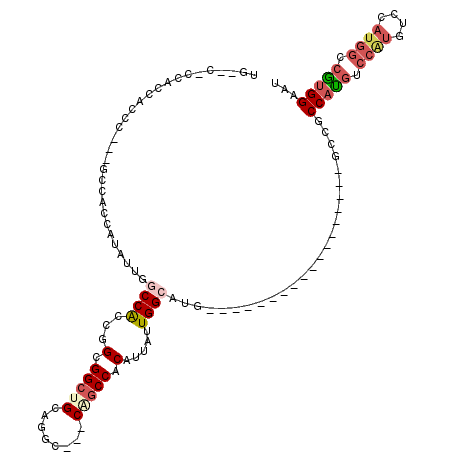
| Location | 12,526,389 – 12,526,485 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.94 |
| Mean single sequence MFE | -38.38 |
| Consensus MFE | -14.96 |
| Energy contribution | -16.48 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.39 |
| SVM decision value | 2.45 |
| SVM RNA-class probability | 0.994097 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12526389 96 + 22224390 UGAACACCACCACCC---GUCCCCAUAUUGGCCACCGGCGGCUGCAGAC---CAGCCACAUUAUUGGCAUG------------------ACCACCAUGUCCAUGUCCAUGGCCAUGGAAU ...............---....(((...((((((..((((((((.....---))))).......(((((((------------------.....)))).))).)))..)))))))))... ( -30.40) >DroSec_CAF1 56002 96 + 1 UGGACACCACCACCC---GGCACCAUAUUGGCCACCGGCGGCUGCAGGC---CCGCCACAUUAUUGGCGUG------------------ACCUCCAUGUCCAUGUCCAUGUCCGUGGAAU ((((((...((..((---((..((.....))...)))).)).((.((((---.(((((......))))).)------------------.))).))))))))..(((((....))))).. ( -33.10) >DroEre_CAF1 56115 89 + 1 -------CUCCAGCC---GCCACCAUAUUGGCCGCCGGCGGCUGGAGGC---CCGCCACAUUAUUGGCAUG------------------GCCGCCAUGUCCAUGGCCAUGGCCGUGGAAC -------.(((((((---(((.((.....)).....))))))))))(((---(.((((......))))..)------------------)))......((((((((....)))))))).. ( -50.00) >DroYak_CAF1 56862 92 + 1 -------CGCCACCA---GCCACCAUAUUGCCCACCGGCGGCUGUAGGC---CGGCCACAUUAUUGGCAUGACC---------------GCCGCCAUGUCCGUGUCCAUGGCCGUGGAAU -------.(((..((---(((.((............)).)))))..)))---..((((......))))....((---------------((.((((((........)))))).))))... ( -37.60) >DroAna_CAF1 66488 114 + 1 UGUCCUCCACCGCCCAGUGGCACCA------CCACCGGCGGCUGCAGGCUGCCACCCACAUUGUUGGCAUGACCACCGACGACUCCACUGCCGCCACUGCCACUGCCACUGCUGGGGAAG .....(((.(((..((((((((...------.((..((((((.((.....))........(((((((........))))))).......))))))..))....)))))))).)))))).. ( -40.80) >consensus UG__C_CCACCACCC___GCCACCAUAUUGGCCACCGGCGGCUGCAGGC___CAGCCACAUUAUUGGCAUG__________________GCCGCCAUGUCCAUGUCCAUGGCCGUGGAAU ..............................((((...(.(((((........))))).).....)))).........................(((((.((((....)))).)))))... (-14.96 = -16.48 + 1.52)


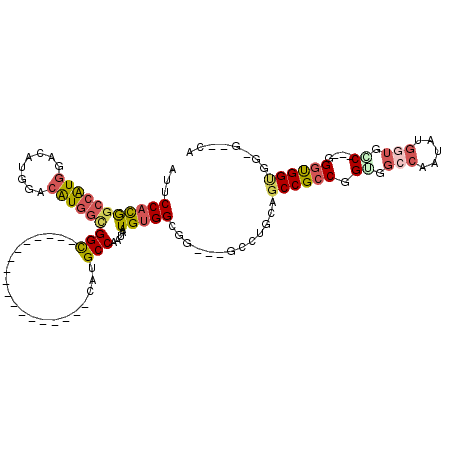
| Location | 12,526,389 – 12,526,485 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.94 |
| Mean single sequence MFE | -47.34 |
| Consensus MFE | -19.62 |
| Energy contribution | -20.78 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.41 |
| SVM decision value | 3.33 |
| SVM RNA-class probability | 0.999024 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12526389 96 - 22224390 AUUCCAUGGCCAUGGACAUGGACAUGGUGGU------------------CAUGCCAAUAAUGUGGCUG---GUCUGCAGCCGCCGGUGGCCAAUAUGGGGAC---GGGUGGUGGUGUUCA ...(((((.((((....)))).))))).(..------------------((.((((......))))))---..).((.((((((.((..((.....))..))---.)))))).))..... ( -37.20) >DroSec_CAF1 56002 96 - 1 AUUCCACGGACAUGGACAUGGACAUGGAGGU------------------CACGCCAAUAAUGUGGCGG---GCCUGCAGCCGCCGGUGGCCAAUAUGGUGCC---GGGUGGUGGUGUCCA ..((((......))))..((((((((.((((------------------(.(((((......))))))---)))).).((((((.(..(((.....)))..)---.)))))).))))))) ( -45.80) >DroEre_CAF1 56115 89 - 1 GUUCCACGGCCAUGGCCAUGGACAUGGCGGC------------------CAUGCCAAUAAUGUGGCGG---GCCUCCAGCCGCCGGCGGCCAAUAUGGUGGC---GGCUGGAG------- ..((((.(((....))).))))......(((------------------(.(((((......))))))---)))(((((((((((.((.......)).))))---))))))).------- ( -50.70) >DroYak_CAF1 56862 92 - 1 AUUCCACGGCCAUGGACACGGACAUGGCGGC---------------GGUCAUGCCAAUAAUGUGGCCG---GCCUACAGCCGCCGGUGGGCAAUAUGGUGGC---UGGUGGCG------- ......((((((((.....((.((((((...---------------.)))))))).....))))))))---(((..(((((((((..........)))))))---))..))).------- ( -45.30) >DroAna_CAF1 66488 114 - 1 CUUCCCCAGCAGUGGCAGUGGCAGUGGCGGCAGUGGAGUCGUCGGUGGUCAUGCCAACAAUGUGGGUGGCAGCCUGCAGCCGCCGGUGG------UGGUGCCACUGGGCGGUGGAGGACA (((((((..((((((((.((.((.(((((((.........((.(((......))).))..((..(((....)))..))))))))).)).------)).))))))))...)).)))))... ( -57.70) >consensus AUUCCACGGCCAUGGACAUGGACAUGGCGGC__________________CAUGCCAAUAAUGUGGCGG___GCCUGCAGCCGCCGGUGGCCAAUAUGGUGCC___GGGUGGUGG_G__CA ...(((((((((((........))))))(((.....................))).....))))).............((((((.((.(((.....))).))....))))))........ (-19.62 = -20.78 + 1.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:33:57 2006