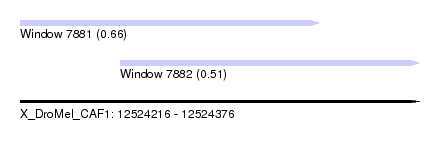
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,524,216 – 12,524,376 |
| Length | 160 |
| Max. P | 0.660700 |

| Location | 12,524,216 – 12,524,336 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.80 |
| Mean single sequence MFE | -48.96 |
| Consensus MFE | -32.84 |
| Energy contribution | -33.28 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.660700 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12524216 120 + 22224390 UUCCCCCACUUACCUAUUAACAUGAUCGGCUGGCGAUGCACGACUGGAGUACUCCGGUCGGCCACGGGGUGCCGAUUCCUUGUCGCCGCCAUAGGCGAAACCACUGGCGCUGCCCACACU ......................((..((((.((((((((.((((((((....))))))))))...(((((....)))))..))))))((((..((.....))..))))))))..)).... ( -41.70) >DroSec_CAF1 53839 116 + 1 ----CCCACUUACCUAUUGGCAUGAUCGGCUGGCGAUGCACGACUGGAGUACUCCGGUCGGCCUCGUGGUGCCGAUUCCUUGUCGCCGCCGUAGGCGAAGCCACUAGCGCUGCCCACACU ----.........(((.((((..((((((((.((((.((.((((((((....)))))))))).)))).).))))))).....(((((......))))).)))).)))............. ( -50.60) >DroEre_CAF1 53930 116 + 1 ----CUCACUCACCUAUUGGCAUGAUCCGCCGGCGAGGCGCGACUGGAGUACUCCGGUCGGCCGCGGGGCGCCGGUUCCUUGUCGCCGCCGUAGGCGAAGCCACUGGCACUGCCCACACU ----............(((((.......)))))((.(((.((((((((....))))))))))).))((((((((((..((..(((((......)))))))..))))))...))))..... ( -52.10) >DroYak_CAF1 54208 116 + 1 ----CACACUCACCUAUUCGCAUGAUCGGCGGGCGAUGCACGACUGGAGUACUCCGGUCGGCCACGUGGCGCCGAUUCCUUGUCGCCGCCAUAGGCAAAGCCACUGGCACUGCCCACACU ----...............((.......))(((((.(((.((((((((....))))))))(((..((((((.((((.....)))).)))))).)))..........))).)))))..... ( -47.80) >DroAna_CAF1 64347 115 + 1 -----ACCCUUACCUGUUGGCGUGUUCGGCCGGGGAGGCCCGGCUGGAGUACUCUGGCCGGCCCCUGGGACCUGGAUCCUUGUCUCCAUAGCCCCCGAAUCCGCCGGCACUGCCCACACU -----.........((((((((..(((((...(((.((((.(((..((....))..))))))))))(((...((((........))))...))))))))..))))))))........... ( -52.60) >consensus ____CACACUUACCUAUUGGCAUGAUCGGCCGGCGAUGCACGACUGGAGUACUCCGGUCGGCCACGGGGCGCCGAUUCCUUGUCGCCGCCGUAGGCGAAGCCACUGGCACUGCCCACACU ...........................(((.(((((.((.((((((((....))))))))))..(((((........))))))))))((((..((.....))..))))...)))...... (-32.84 = -33.28 + 0.44)


| Location | 12,524,256 – 12,524,376 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.17 |
| Mean single sequence MFE | -55.68 |
| Consensus MFE | -43.44 |
| Energy contribution | -43.20 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.78 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.513679 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12524256 120 + 22224390 CGACUGGAGUACUCCGGUCGGCCACGGGGUGCCGAUUCCUUGUCGCCGCCAUAGGCGAAACCACUGGCGCUGCCCACACUGUUUCGUGGCGAUGGUGACUCCUUGUCGGAACCACCGCCG .(((((((....)))))))(((...(((((.(((((.....((((((((((..((.....))..))))..((((.((........))))))..)))))).....))))).))).))))). ( -52.90) >DroSec_CAF1 53875 120 + 1 CGACUGGAGUACUCCGGUCGGCCUCGUGGUGCCGAUUCCUUGUCGCCGCCGUAGGCGAAGCCACUAGCGCUGCCCACACUGUUUCGUGGCGAGGGCGACUCCUUGUCGGAACCACCACCG ((((((((....)))))))).....(((((.(((((.....(((((((((...)))...(((((.((((.((....)).))))..)))))...)))))).....))))).)))))..... ( -55.20) >DroEre_CAF1 53966 120 + 1 CGACUGGAGUACUCCGGUCGGCCGCGGGGCGCCGGUUCCUUGUCGCCGCCGUAGGCGAAGCCACUGGCACUGCCCACACUGUUGCGCGGCGAGGGCGACUCCUUGUCGGAGCCACCGCCG ((((((((....))))))))((((((((((((((((..((..(((((......)))))))..))))))...)))).((....))))))))...((((.((((.....))))....)))). ( -60.90) >DroYak_CAF1 54244 120 + 1 CGACUGGAGUACUCCGGUCGGCCACGUGGCGCCGAUUCCUUGUCGCCGCCAUAGGCAAAGCCACUGGCACUGCCCACACUGUUGCGCGGCGAGGGCGACUCCUUGUCGGAGCCACCGCCG .(((((((....)))))))(((...(((((.(((((.....((((((((((..(((...)))..)))).((((((.((....)).).))).)))))))).....))))).))))).))). ( -62.00) >DroAna_CAF1 64382 120 + 1 CGGCUGGAGUACUCUGGCCGGCCCCUGGGACCUGGAUCCUUGUCUCCAUAGCCCCCGAAUCCGCCGGCACUGCCCACACUGUUCCGGGGCGAGGGCGACUCCUUGUCAGAGCCACCGCCU .(((..((....))..)))(((.....((..((((......(((.((...(((((.((((.....(((...)))......)))).)))))...)).)))......))))..))...))). ( -47.40) >consensus CGACUGGAGUACUCCGGUCGGCCACGGGGCGCCGAUUCCUUGUCGCCGCCGUAGGCGAAGCCACUGGCACUGCCCACACUGUUGCGCGGCGAGGGCGACUCCUUGUCGGAGCCACCGCCG .(((((((....)))))))(((.....(((.(((((.....((((((((((..((.....))..))))..(((((..........).))))..)))))).....))))).)))...))). (-43.44 = -43.20 + -0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:33:54 2006