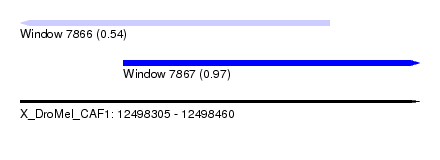
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,498,305 – 12,498,460 |
| Length | 155 |
| Max. P | 0.973670 |

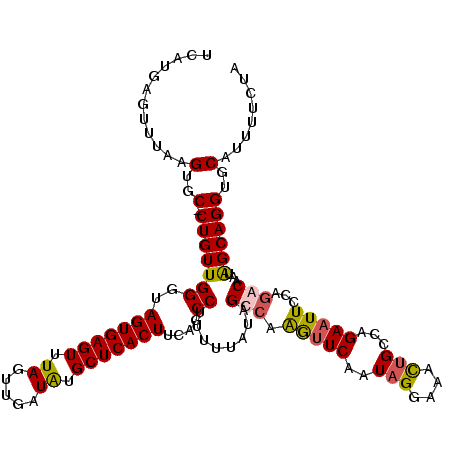
| Location | 12,498,305 – 12,498,425 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.26 |
| Mean single sequence MFE | -33.77 |
| Consensus MFE | -23.29 |
| Energy contribution | -23.47 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.535351 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12498305 120 - 22224390 UCAUGAGUUUAAGUGCCCUGUUGGGUAGUGAGUUUAGUUGAUAUGCUCACUUUAGCCUUUUUAUAGCCAGGUUCAAUAGGAAACUGCCAGAAUUCUAGGCAACAGCAGGUGCAUGUUCUA ............(((((((((((((((((((((.((.....)).)))))))...)))........(((((((((..(((....)))...))).))).)))..))))))).))))...... ( -39.40) >DroSec_CAF1 27782 120 - 1 UCAUGAGUUUAAGUGCCCUGUUGGAUAGUGAGUUUAGUUGAUAUGCUCACUUUAGCCUUUUUAUAGCCAGGUUCAAUAGGAAACUGCCAGAAUUCCAGGCAGUAGCAGGUGCAUAUUCCA ....((((....((((((((((((..(((((((.((.....)).)))))))....))........(((.(((((..(((....)))...)))..)).)))...)))))).)))))))).. ( -35.20) >DroEre_CAF1 28873 119 - 1 UCAUGAGUUUAAGUGC-CUGUUGGGUAGUGAGUUUAUUUGAUGUGCUCACUUCAGCCAUUUUAUAGCCAAAUUCAAUAGGAAACUGCCAGAAUUCUACACAAUGGCAGGUGCAUUUUCUA ............(..(-((....((.(((((((...........))))))))).((((((.(.(((...(((((..(((....)))...)))))))).).)))))))))..)........ ( -30.60) >DroYak_CAF1 23237 119 - 1 UCAUGAGUUUAAGUGC-CUGUUGGUUAGUGAGUUUAGUUGAUGUGCUCACUUCAGCCUUUUUAUAGCCAAGCUCAAUAGAAAAUGGCCAGAAUUCCAGACAAUAGCAGGUGCAUUUUCUA ............(..(-((((((((((((((((.((.....)).)))))))..))))........((((...((....))...))))................))))))..)........ ( -29.90) >consensus UCAUGAGUUUAAGUGC_CUGUUGGGUAGUGAGUUUAGUUGAUAUGCUCACUUCAGCCUUUUUAUAGCCAAGUUCAAUAGGAAACUGCCAGAAUUCCAGACAAUAGCAGGUGCAUUUUCUA ............(..(.(((((((..(((((((.((.....)).)))))))....))........(((.(((((..(((....)))...)))))...)))...))))))..)........ (-23.29 = -23.47 + 0.19)


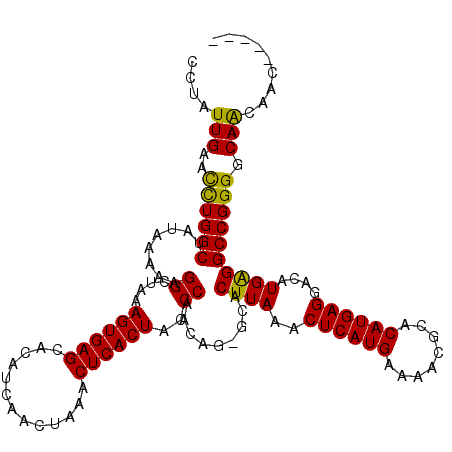
| Location | 12,498,345 – 12,498,460 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.48 |
| Mean single sequence MFE | -30.42 |
| Consensus MFE | -26.02 |
| Energy contribution | -25.14 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.973670 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12498345 115 + 22224390 CCUAUUGAACCUGGCUAUAAAAAGGCUAAAGUGAGCAUAUCAACUAAACUCACUACCCAACAGGGCACUUAAACUCAUGAAAACGCACAUGAGGACAUGAGGCCGGGGCAGCAAC----- ....(((..(((((((.....(((.....((((((.............)))))).(((....)))..)))...((((((........)))))).......)))))))....))).----- ( -30.92) >DroSec_CAF1 27822 115 + 1 CCUAUUGAACCUGGCUAUAAAAAGGCUAAAGUGAGCAUAUCAACUAAACUCACUAUCCAACAGGGCACUUAAACUCAUGAAAACGCACAUGAGGACAUGAGGCCGGGGCAGCAGC----- ....(((..(((((((.....(((((...((((((.............))))))..((....)))).)))...((((((........)))))).......)))))))....))).----- ( -30.12) >DroEre_CAF1 28913 119 + 1 CCUAUUGAAUUUGGCUAUAAAAUGGCUGAAGUGAGCACAUCAAAUAAACUCACUACCCAACAG-GCACUUAAACUCAUGAAAACGCACAUGAGGACAUGGGGCCGGGGCAACAACAGCUA ......................((((((.((((((.............)))))).(((....(-((.((((..((((((........))))))....))))))).)))......)))))) ( -32.12) >DroYak_CAF1 23277 119 + 1 UCUAUUGAGCUUGGCUAUAAAAAGGCUGAAGUGAGCACAUCAACUAAACUCACUAACCAACAG-GCACUUAAACUCAUGAAAACGCACAUGAGGACAUGAGGCCGGGGCAACAACAACAA ........(((..(((.......)))..)((((((.............))))))..((....(-((.((((..((((((........))))))....))))))).))))........... ( -28.52) >consensus CCUAUUGAACCUGGCUAUAAAAAGGCUAAAGUGAGCACAUCAACUAAACUCACUACCCAACAG_GCACUUAAACUCAUGAAAACGCACAUGAGGACAUGAGGCCGGGGCAACAAC_____ ....(((..((((((........((....((((((.............))))))..)).........((((..((((((........))))))....)))))))))).)))......... (-26.02 = -25.14 + -0.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:33:39 2006