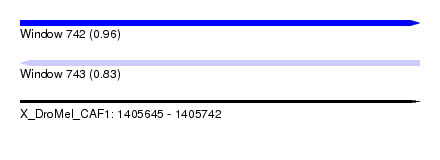
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,405,645 – 1,405,742 |
| Length | 97 |
| Max. P | 0.961942 |

| Location | 1,405,645 – 1,405,742 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 87.74 |
| Mean single sequence MFE | -23.16 |
| Consensus MFE | -16.97 |
| Energy contribution | -17.84 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961942 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1405645 97 + 22224390 CACAUUCACAGCCACAGCCACAUCCACAGCCAUAUCCAC------------AUCCACGGCUGUGUGUGUGCAUCUUUUUGCUUUCGUUUUAGAAAACUCACGGCUCGCA ..........((...(((((((..(((((((........------------......)))))))..)))(((......)))((((......))))......)))).)). ( -26.64) >DroSec_CAF1 11073 103 + 1 CACAUCUACAGCCACAUCCACAUUCACAGCCAUAUCCAC------AUCCACAUCCACGGCUGUGUGUGUGCAUCUUUUUGCUUUCGUUUUAGAAAACUCACGGUUCGCA .........((((.....(((((.(((((((........------............))))))).)))))...........((((......))))......)))).... ( -21.95) >DroSim_CAF1 8273 103 + 1 CACAUCCACGGCCACAUCCACAUUCACAGCCAUAUCCAC------AUCCACAUCCACGGCUGUGUGUGUGCAUCUUUUUGCUUUCGUUUUAGAAAACUCACGGCUCGCA ........(((((.....(((((.(((((((........------............))))))).)))))...........((((......))))......))).)).. ( -25.15) >DroEre_CAF1 7507 107 + 1 CAUAUCCACAGCAACAUCCACAUCCACAUCCACAUCCACGUCCACGUCCACAUCCACAGCU--GUGUGUGCAUCUUUUUGCUUUCGUUUUAGAAAACUCACGGCUCGCA ..........((.........................(((....)))..........((((--(((.(((((......)))((((......)))))).))))))).)). ( -18.90) >consensus CACAUCCACAGCCACAUCCACAUCCACAGCCAUAUCCAC______AUCCACAUCCACGGCUGUGUGUGUGCAUCUUUUUGCUUUCGUUUUAGAAAACUCACGGCUCGCA .........((((.....(((((.(((((((..........................))))))).)))))...........((((......))))......)))).... (-16.97 = -17.84 + 0.88)

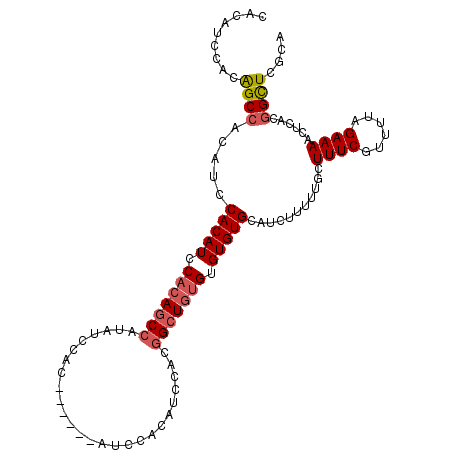
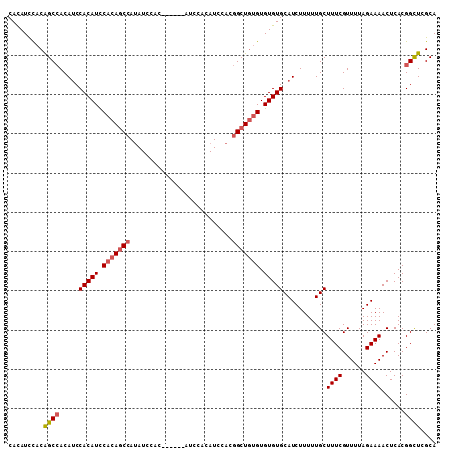
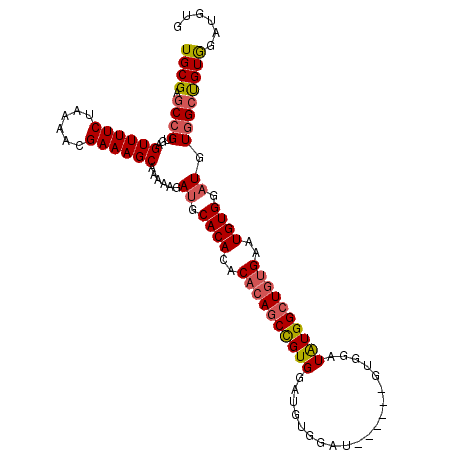
| Location | 1,405,645 – 1,405,742 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 87.74 |
| Mean single sequence MFE | -30.50 |
| Consensus MFE | -24.12 |
| Energy contribution | -24.88 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.829573 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1405645 97 - 22224390 UGCGAGCCGUGAGUUUUCUAAAACGAAAGCAAAAAGAUGCACACACACAGCCGUGGAU------------GUGGAUAUGGCUGUGGAUGUGGCUGUGGCUGUGAAUGUG .((.(((((...((((((......)))))).........((((..((((((((((...------------.....))))))))))..))))....)))))))....... ( -34.60) >DroSec_CAF1 11073 103 - 1 UGCGAACCGUGAGUUUUCUAAAACGAAAGCAAAAAGAUGCACACACACAGCCGUGGAUGUGGAU------GUGGAUAUGGCUGUGAAUGUGGAUGUGGCUGUAGAUGUG ((((..((....((((((......))))))....(.((.((((..((((((((((..(......------..)..))))))))))..)))).)).))).))))...... ( -29.00) >DroSim_CAF1 8273 103 - 1 UGCGAGCCGUGAGUUUUCUAAAACGAAAGCAAAAAGAUGCACACACACAGCCGUGGAUGUGGAU------GUGGAUAUGGCUGUGAAUGUGGAUGUGGCCGUGGAUGUG .(((.(((....((((((......))))))....(.((.((((..((((((((((..(......------..)..))))))))))..)))).)).)))))))....... ( -35.10) >DroEre_CAF1 7507 107 - 1 UGCGAGCCGUGAGUUUUCUAAAACGAAAGCAAAAAGAUGCACACAC--AGCUGUGGAUGUGGACGUGGACGUGGAUGUGGAUGUGGAUGUGGAUGUUGCUGUGGAUAUG .((((.((....((((((......))))))....(.((.(((((((--(.(..((.(((....)))...))..).))))..)))).)).)))...)))).......... ( -23.30) >consensus UGCGAGCCGUGAGUUUUCUAAAACGAAAGCAAAAAGAUGCACACACACAGCCGUGGAUGUGGAU______GUGGAUAUGGCUGUGAAUGUGGAUGUGGCUGUGGAUGUG ((((.((((...((((((......))))))......((.((((..((((((((((....................))))))))))..)))).)).))))))))...... (-24.12 = -24.88 + 0.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:45:39 2006