
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,489,792 – 12,489,912 |
| Length | 120 |
| Max. P | 0.717409 |

| Location | 12,489,792 – 12,489,912 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.06 |
| Mean single sequence MFE | -43.30 |
| Consensus MFE | -27.52 |
| Energy contribution | -27.75 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.64 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.513629 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
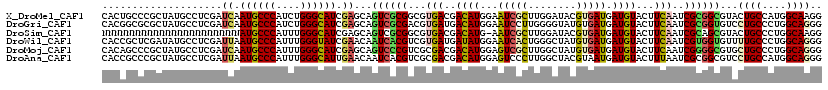
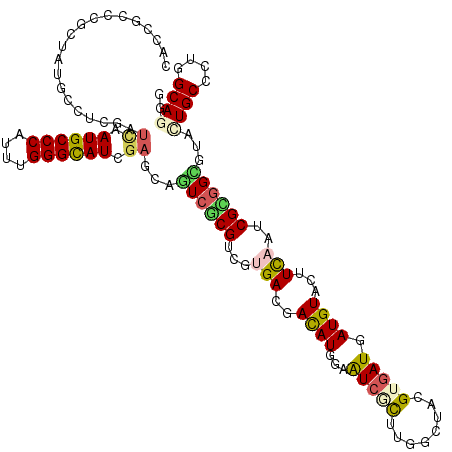
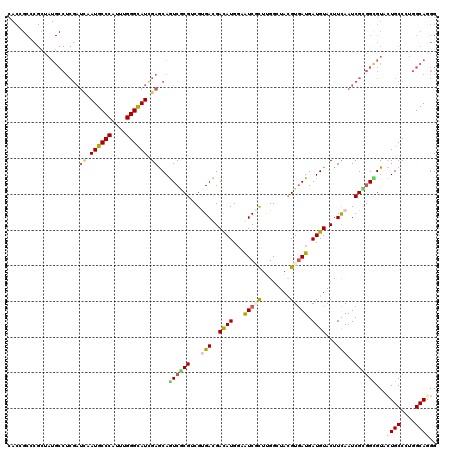
>X_DroMel_CAF1 12489792 120 + 22224390 CACUGCCCGCUAUGCCUCGAUCAAUGCCCAUCUGGGCAUCGAGCAGUCGCGGCGUGACGACAUGGAAUCGCUUGGAUACGUGAUGAUGUACUUCAAUCGCGGCGUACUGCCAUGGCAAGG ........((((((......((.((((((....)))))).))((((((((.((((((.(((((...(((((........))))).)))).).)))..))).))).))))))))))).... ( -45.80) >DroGri_CAF1 23417 120 + 1 CACGGCGCGCUAUGCCUCGAUCAAUGCCCAUCUGGGCAUCGAGCAGUCGCGACGUGAUGACAUGGAAUCCUUGGGGUAUGUGAUGAUGUACUUCAAUCGCGGUGUCCUGCCCUGGCAGGG ((((.((((...(((.((((....(((((....))))))))))))..)))).))))..(((((.(.....(((((((((((....)))))))))))...).)))))((((....)))).. ( -50.10) >DroSim_CAF1 20822 119 + 1 NNNNNNNNNNNNNNNNNNNNNNNAUGCCCAUUUGGGCAUCGAGCAGUCGCGGCGUGACGACAUG-AAUCGCUUGGAUACGUGAUGAUGUACUUCAAUCGCAGCGUACUGCCCUGGCAAGG .......................((((((....))))))...((((((((.((((((.(((((.-.(((((........))))).)))).).)))..))).))).)))))(((....))) ( -36.10) >DroWil_CAF1 32209 120 + 1 CACCGCUCGAUAUGCCUCGAUUAAUGCCCAUUUGGGUAUCGAACAAUCACGUCGUGAUGAUAUGGAAUCACUGGGCUAUGUGAUGAUGUACUUCAAUCGUGGUGUUUUGCCCUGGCAGGG ..((..((((......))))....((((.....(((((..(((((.(((((...(((.(.((((..(((((........)))))..))))).)))..))))))))))))))).)))))). ( -36.10) >DroMoj_CAF1 25467 120 + 1 CACAGCCCGCUAUGCCUCGAUCAAUGCCCAUUUGGGCAUCGAGCAGUCCCGUCGCGACGACAUGGAGUCGCUUGGCUAUGUGAUGAUGUACUUCAAUCGGGGCGUGCUGCCCUGGCAGGG .....(((((.(((((((((((.((((((....)))))).))(((...(((..(((((........))))).)))...)))..(((......))).)))))))))))(((....)))))) ( -48.60) >DroAna_CAF1 26079 120 + 1 CACCGCCCGCUAUGCCUCGAUUAAUGCCCAUUUGGGCAUUGAACAAUCACGUCGCGACGACAUGGAGUCCCUUGGCUACGUAAUGAUGUACUUUAAUCGCGGCGUCCUGCCAUGGCAGGG ...........(((((.((((((((((((....)))))))))......((((((..(((.((.((....)).))....)))..)))))).......))).)))))(((((....))))). ( -43.10) >consensus CACCGCCCGCUAUGCCUCGAUCAAUGCCCAUUUGGGCAUCGAGCAGUCGCGUCGUGACGACAUGGAAUCGCUUGGCUACGUGAUGAUGUACUUCAAUCGCGGCGUACUGCCCUGGCAGGG ....................((.((((((....)))))).))...((((((...(((..((((...(((((........))))).))))...)))..))))))...((((....)))).. (-27.52 = -27.75 + 0.23)
| Location | 12,489,792 – 12,489,912 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.06 |
| Mean single sequence MFE | -41.42 |
| Consensus MFE | -27.49 |
| Energy contribution | -26.83 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.717409 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
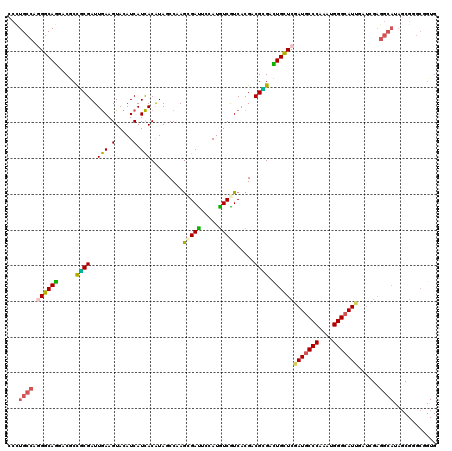
>X_DroMel_CAF1 12489792 120 - 22224390 CCUUGCCAUGGCAGUACGCCGCGAUUGAAGUACAUCAUCACGUAUCCAAGCGAUUCCAUGUCGUCACGCCGCGACUGCUCGAUGCCCAGAUGGGCAUUGAUCGAGGCAUAGCGGGCAGUG ((.((((...(((((.(((.(((.(((..((((........)))).)))(((((.....)))))..))).))))))))(((((((((....)))))))))....))))....))...... ( -45.00) >DroGri_CAF1 23417 120 - 1 CCCUGCCAGGGCAGGACACCGCGAUUGAAGUACAUCAUCACAUACCCCAAGGAUUCCAUGUCAUCACGUCGCGACUGCUCGAUGCCCAGAUGGGCAUUGAUCGAGGCAUAGCGCGCCGUG .(((((....))))).((((((((((((.(.((((.(((............)))...))))).))).))))))..((.(((((((((....))))))))).)).(((.......)))))) ( -41.60) >DroSim_CAF1 20822 119 - 1 CCUUGCCAGGGCAGUACGCUGCGAUUGAAGUACAUCAUCACGUAUCCAAGCGAUU-CAUGUCGUCACGCCGCGACUGCUCGAUGCCCAAAUGGGCAUNNNNNNNNNNNNNNNNNNNNNNN ........(((((((.(((.(((.(((..((((........)))).)))(((((.-...)))))..))).)))))))))).((((((....))))))....................... ( -36.50) >DroWil_CAF1 32209 120 - 1 CCCUGCCAGGGCAAAACACCACGAUUGAAGUACAUCAUCACAUAGCCCAGUGAUUCCAUAUCAUCACGACGUGAUUGUUCGAUACCCAAAUGGGCAUUAAUCGAGGCAUAUCGAGCGGUG ...(((....)))...((((.(((((((.((........))...((((((((((........)))))((((....))))...........))))).)))))))..((.......)))))) ( -29.40) >DroMoj_CAF1 25467 120 - 1 CCCUGCCAGGGCAGCACGCCCCGAUUGAAGUACAUCAUCACAUAGCCAAGCGACUCCAUGUCGUCGCGACGGGACUGCUCGAUGCCCAAAUGGGCAUUGAUCGAGGCAUAGCGGGCUGUG ..((((....))))(((((((((((.((......))))).....(((..(((..(((.(((((...)))))))).)))(((((((((....)))))))))....)))...).)))).))) ( -46.40) >DroAna_CAF1 26079 120 - 1 CCCUGCCAUGGCAGGACGCCGCGAUUAAAGUACAUCAUUACGUAGCCAAGGGACUCCAUGUCGUCGCGACGUGAUUGUUCAAUGCCCAAAUGGGCAUUAAUCGAGGCAUAGCGGGCGGUG .(((((....))))).(((((((((........)))....(((.(((...((((..(((((((...)))))))...))))(((((((....)))))))......)))...))).)))))) ( -49.60) >consensus CCCUGCCAGGGCAGGACGCCGCGAUUGAAGUACAUCAUCACAUAGCCAAGCGAUUCCAUGUCGUCACGACGCGACUGCUCGAUGCCCAAAUGGGCAUUGAUCGAGGCAUAGCGGGCGGUG ...((((.((((((.....((((..(((.(.....).))).........(((((.....))))).....)))).))))))(((((((....)))))))......))))............ (-27.49 = -26.83 + -0.66)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:33:30 2006