
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,485,023 – 12,485,131 |
| Length | 108 |
| Max. P | 0.778540 |

| Location | 12,485,023 – 12,485,131 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.83 |
| Mean single sequence MFE | -21.92 |
| Consensus MFE | -17.59 |
| Energy contribution | -17.26 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.778540 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12485023 108 + 22224390 ------AACCAAAC---CCGAA---UAAAUUACAGAUAAAUUGGAUGGAACUUUCUCAAGAUCCAGAUCAUCGUCAAUGUCCAGCAUUGAUAUGUCUUCCUCUGAAUCUGUUACCUGUUU ------........---..(((---((....((((((.....(((((((.(((....))).))))((.(((.(((((((.....)))))))))))).))).....))))))....))))) ( -24.20) >DroVir_CAF1 19680 113 + 1 CACACACAC-------AUAAAAUACUGACUUGCAGAUAAAUUGGAUGGAACCUUCUCGAGAUCCAGAUCGUCAUCAAUGUCCAGCAUUGAUAUGUCUUCCUCUGAAUCUGUUACCUGUUU .........-------..........(((..((((((...(..((.((((.......(((((....))).))(((((((.....))))))).....))))))..))))))).....))). ( -21.20) >DroGri_CAF1 16725 103 + 1 ------UAC-----------AAAACUGCCUUGCAGAUAAAUUGGAUGGUACCUUCUCGAGAUCCAGAUCGUCAUCAAUGUCCAGCAUUGAUAUGUCUUCCUCUGAAUCUGUUACCUGCCU ------...-----------....((((...)))).......((..((((.(...(((((....(((.(((.(((((((.....)))))))))))))..))).))....).))))..)). ( -20.90) >DroWil_CAF1 27413 101 + 1 ------AAAUAAUC---C----------CUUACAGAUAAAUUGGAUGGCACUUUUUCAAGAUCCAGAUCAUCUUCAAUGUCCAGCAUUGAUAUGUCUUCCUCGGAAUCUGUUACUUGUCU ------........---.----------...((((((.....(((.((((.........(((....)))....((((((.....))))))..)))).))).....))))))......... ( -20.10) >DroMoj_CAF1 17691 104 + 1 ------UAC----------UAAAACUGACUUGCAGAUAAAUUAGAUGGCACCUUCUCGAGAUCCAGAUCGUCAUCAAUGUCGAGCAUUGAUAUGUCUUCCUCUGAAUCUGUUACCUGUUU ------...----------..((((......((((((...(((((.((((.......(((((....))).))(((((((.....))))))).))))....))))))))))).....)))) ( -21.30) >DroAna_CAF1 18114 111 + 1 ------AAAAAAACCAAUCGAA---UAAAUUACAGAUAAAUUGGAUGGAACUUUCUCCAGAUCCAGAUCAUCGUCAAUGUCCAGCAUUGAUAUGUCUUCCUCUGAAUCUGUUACCUGUUU ------.............(((---((....((((((....((((((((......))))..))))((.(((.(((((((.....)))))))))))).........))))))....))))) ( -23.80) >consensus ______AAC___________AA___UGACUUACAGAUAAAUUGGAUGGAACCUUCUCGAGAUCCAGAUCAUCAUCAAUGUCCAGCAUUGAUAUGUCUUCCUCUGAAUCUGUUACCUGUUU ...............................((((((.....((((((..............)))((.(((.(((((((.....)))))))))))).))).....))))))......... (-17.59 = -17.26 + -0.33)
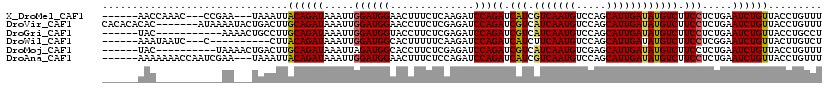

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:33:26 2006