
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,448,484 – 12,448,595 |
| Length | 111 |
| Max. P | 0.745954 |

| Location | 12,448,484 – 12,448,595 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 69.85 |
| Mean single sequence MFE | -33.23 |
| Consensus MFE | -12.05 |
| Energy contribution | -12.38 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.36 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.745954 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
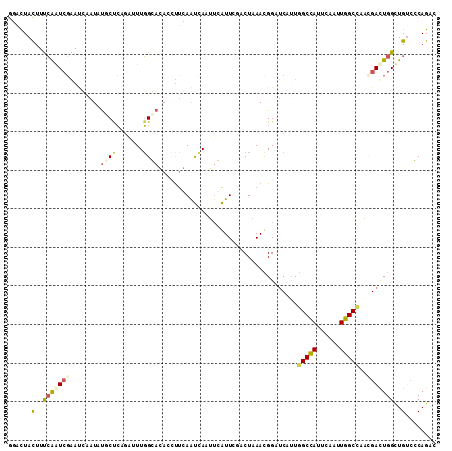
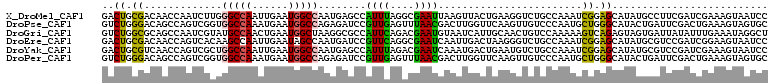
>X_DroMel_CAF1 12448484 111 + 22224390 GGAUUACUUUCGAUCGAAGGCAUAUGCUCCGAUUUGGCAGACCUUCAGUAACUUAAUUCGCCUAAAUGGCUCAUUGGCCAUUCAAUUGGCCCAAGAUUGGUUGUCGCAGUC .((((((..(((((((((((....((((.......))))..))))).............(((.....))).....(((((......)))))...))))))..))...)))) ( -29.90) >DroPse_CAF1 1745 111 + 1 GCACUACUUUCAGUCGAAUCAGUAUGCCCAGCAUUGGGACAACUUGAACCAAGUCGUUAAACUCAACGGAUCUCUGGCCAUUCAUUUGGCCACCGACUGGCUGUCCCAGAC (((.(((((((....)))..)))))))......(((((((((((((...)))))............(((.((..((((((......))))))..)))))..)))))))).. ( -32.70) >DroGri_CAF1 1494 111 + 1 AGCCUAUUUCAAAUAUAAUCACUACUCUGACUUUUUGGACAGUUGCAAUGAUUACAUUCGUCUGAAUGGCGCCUUAGCCAUUCAGUUGGCAUACGAUUGGCUGCGCCAGAC .(((...........((((((.....(((.((....)).)))......)))))).......(((((((((......)))))))))..)))......(((((...))))).. ( -30.10) >DroEre_CAF1 1823 111 + 1 GGAUUACUUCCGAUCGGACGCAUAUGCUCCGAUUUGGCAGACCCUUAGUCAAUUGAUUCGCCUGAACGGAUCAUUGGCUAUUCAAUUGGCUUGUGACUGGUUGUCGCAGUC (((.....)))(((((((.((....))))))))).(.(((.....((((((((.((((((......)))))))))))))).....))).)(((((((.....))))))).. ( -38.40) >DroYak_CAF1 1524 111 + 1 GGAUUACUUUCGAUCGGACGCAUAUGCUCCGAUUUGGCAGACAUUCAGUCAUUUGAUUCGUCUAAAUGGCUCAUUGGCCAUUCAAUUGGCCAGCGACUGGUUGACGCAGUC ((((..((.(((((((((.((....))))))).)))).))..))))((((((((((....)).))))))))..(((((((......))))))).((((((....).))))) ( -35.60) >DroPer_CAF1 1745 111 + 1 GCACUACUUUCAGUCGAAUCAGUAUGCCCAGCAUUGGGACAACUUGAACCAAGUCGUUAAACUCAACGGAUCUCUGGCCAUUCAUUUGGCCACCGACUGGCUGUCCCAGAC (((.(((((((....)))..)))))))......(((((((((((((...)))))............(((.((..((((((......))))))..)))))..)))))))).. ( -32.70) >consensus GGACUACUUUCAAUCGAAUCAAUAUGCUCAGAUUUGGCACACCUUCAAUCAAUUCAUUCGACUAAACGGAUCAUUGGCCAUUCAAUUGGCCAACGACUGGCUGUCCCAGAC ......(..(((((((........((((.......))))....................................(((((......)))))..)))))))..)........ (-12.05 = -12.38 + 0.34)


| Location | 12,448,484 – 12,448,595 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 69.85 |
| Mean single sequence MFE | -34.95 |
| Consensus MFE | -9.52 |
| Energy contribution | -8.00 |
| Covariance contribution | -1.52 |
| Combinations/Pair | 1.54 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.27 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.571809 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12448484 111 - 22224390 GACUGCGACAACCAAUCUUGGGCCAAUUGAAUGGCCAAUGAGCCAUUUAGGCGAAUUAAGUUACUGAAGGUCUGCCAAAUCGGAGCAUAUGCCUUCGAUCGAAAGUAAUCC ((.((((((..(((....)))(((...((((((((......))))))))))).......)))..(((((((.((((......).)))...))))))).......))).)). ( -28.80) >DroPse_CAF1 1745 111 - 1 GUCUGGGACAGCCAGUCGGUGGCCAAAUGAAUGGCCAGAGAUCCGUUGAGUUUAACGACUUGGUUCAAGUUGUCCCAAUGCUGGGCAUACUGAUUCGACUGAAAGUAGUGC ((((((((((((..(((..((((((......))))))..)))...(..((((....))))..).....)))))))))((((...))))........)))............ ( -35.20) >DroGri_CAF1 1494 111 - 1 GUCUGGCGCAGCCAAUCGUAUGCCAACUGAAUGGCUAAGGCGCCAUUCAGACGAAUGUAAUCAUUGCAACUGUCCAAAAAGUCAGAGUAGUGAUUAUAUUUGAAAUAGGCU (..((((...))))..)....(((..(((((((((......))))))))).((((((((((((((((..(((.(......).))).)))))))))))))))).....))). ( -44.00) >DroEre_CAF1 1823 111 - 1 GACUGCGACAACCAGUCACAAGCCAAUUGAAUAGCCAAUGAUCCGUUCAGGCGAAUCAAUUGACUAAGGGUCUGCCAAAUCGGAGCAUAUGCGUCCGAUCGGAAGUAAUCC (((((.(....))))))...((.(((((((...(((..(((.....))))))...))))))).))..((((((.((..((((((((....)).)))))).)).))..)))) ( -32.80) >DroYak_CAF1 1524 111 - 1 GACUGCGUCAACCAGUCGCUGGCCAAUUGAAUGGCCAAUGAGCCAUUUAGACGAAUCAAAUGACUGAAUGUCUGCCAAAUCGGAGCAUAUGCGUCCGAUCGAAAGUAAUCC (((((.(....))))))..((((.....(((((((......)))))))(((((..(((......))).))))))))).((((((((....)).))))))(....)...... ( -33.70) >DroPer_CAF1 1745 111 - 1 GUCUGGGACAGCCAGUCGGUGGCCAAAUGAAUGGCCAGAGAUCCGUUGAGUUUAACGACUUGGUUCAAGUUGUCCCAAUGCUGGGCAUACUGAUUCGACUGAAAGUAGUGC ((((((((((((..(((..((((((......))))))..)))...(..((((....))))..).....)))))))))((((...))))........)))............ ( -35.20) >consensus GACUGCGACAACCAGUCGCUGGCCAAUUGAAUGGCCAAUGAGCCAUUCAGACGAAUGAAUUGACUGAAGGUCUCCCAAAUCGGAGCAUAUGCAUUCGAUCGAAAGUAAUCC ..((.((.............(((((......)))))........((((....))))........................)).)).......................... ( -9.52 = -8.00 + -1.52)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:33:12 2006