
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,447,933 – 12,448,045 |
| Length | 112 |
| Max. P | 0.526234 |

| Location | 12,447,933 – 12,448,045 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 77.79 |
| Mean single sequence MFE | -28.12 |
| Consensus MFE | -17.99 |
| Energy contribution | -19.02 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.64 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.526234 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12447933 112 + 22224390 --GAUGAUGAAU-ACGUCUUC----AUAUAGGGCUCCACCUGCGCCUUGACGGAAUCUCUGCUUCGCCAUCAAGGUUUCCGCACGGGAUUCUUCAGUUCUCCCCACAUCCUCUUCACGA --((.((((...-.((((...----...((((......))))......))))((((((((((...(((.....)))....))).)))))))..............)))).))....... ( -28.30) >DroPse_CAF1 1195 111 + 1 ----UAAUG-AC-ACCUCCUC--CUUUGCAGGGCUCAACCUGUGCUCUCACGGAAUCCCUGCUGCGUCACAACGGAGUUCACACUGGAUUCUUCAGCUCUCCGCACAUUGUGUACACCA ----.....-..-........--....((((((......(((((....)))))...))))))...((((((((((((......(((((...)))))..)))))....))))).)).... ( -27.60) >DroSec_CAF1 941 112 + 1 --GAUGAUGAAU-ACGUCUUC----ACAUAGGGCUCCACCUGCGCCUUGACGGAAUCUCUGCUUCGCCAUCAAGGUGUCCGCACGGGAUUCUUCAGCUCCCCCCACAUCCUGUUCACCA --..((.(((((-(((((...----.(.((((......)))).)....))))((((((((((..((((.....))))...))).)))))))...................)))))).)) ( -30.50) >DroSim_CAF1 939 112 + 1 --GAUGAUGAAU-ACGUCUUC----AUAUAGGGCUCCACCUGCGCCUUGACGGAAUCUCUGCUUCGCCAUCAAGGUGUCCGCACGGGAUUCUUCAGCUCCCCCCACAUCCUGUUCACCA --..((.(((((-(((((...----...((((......))))......))))((((((((((..((((.....))))...))).)))))))...................)))))).)) ( -30.50) >DroYak_CAF1 966 119 + 1 AUACUAAUGAUCCAUGACUUCACUUACAUAGGGCUCCACCUGCGCCUUGACGGAAUCUCUGCUUCGCCAUCAGGGUGUCCGCACGGGUUUCUUCAGUUCCCCCCACAUUCUGUCCACCA ...............(((...........(((((.......)).)))(((.(((((((.(((..((((.....))))...))).))))))).)))................)))..... ( -24.00) >DroAna_CAF1 1627 101 + 1 -----------U-UCGUC--C----UUUUAGGGCUCUACCUGCGCCUUGACGGAGUCCUUGCUGCGUCAUCAUGGCUACCGGACUGGCUUCUUCAGUUCACCCCACAUCCUGUCCACCA -----------.-.....--.----.....((((.........(((.(((((.(((....))).)))))....)))....(((((((.....)))))))............)))).... ( -27.80) >consensus __GAUGAUGAAU_ACGUCUUC____AUAUAGGGCUCCACCUGCGCCUUGACGGAAUCUCUGCUUCGCCAUCAAGGUGUCCGCACGGGAUUCUUCAGCUCCCCCCACAUCCUGUUCACCA .....((((...................((((((.(.....).))))))..(((((((((((..((((.....))))...))).)))))))).............)))).......... (-17.99 = -19.02 + 1.03)
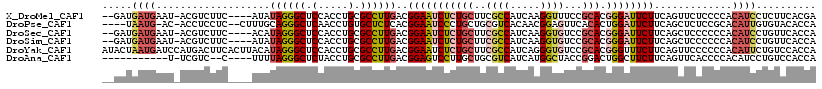
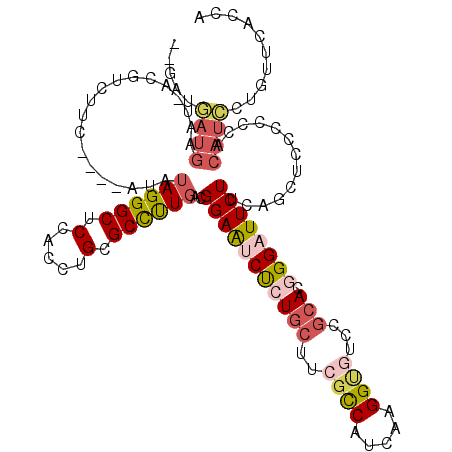
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:33:10 2006