
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,432,504 – 12,432,637 |
| Length | 133 |
| Max. P | 0.912951 |

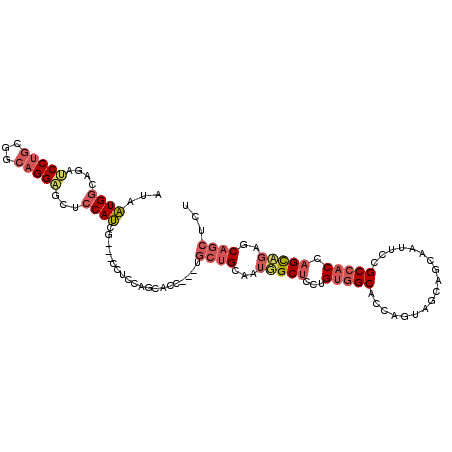
| Location | 12,432,504 – 12,432,610 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 72.53 |
| Mean single sequence MFE | -35.85 |
| Consensus MFE | -15.88 |
| Energy contribution | -17.29 |
| Covariance contribution | 1.41 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.783556 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12432504 106 + 22224390 AUAAUGGCAGAUCCUGCAGCAGGAGCUCCAUCG---CCUCCAGCUCCUGCUGCUGCAAUUGCUCCUGUGGCACCAGUAGCACCAAUUCCGCCACCAGCAGAUCAGCUCU .....(((.((((..((((((((((((......---.....))))))))))))......((((...(((((..................))))).)))))))).))).. ( -39.77) >DroSec_CAF1 5889 106 + 1 AUAAUGACAGACCCUGCGGCAGGGGCUCCAUCG---CCUCCAGCUCCUGUUGCUGCAAUUGCUCCUGUGGCACCAGUCGCAGCAAUUCCGCCACCAGCAGAACAGCUCU .............((((((((((((((......---.....))))))))))((.(.(((((((...(((((....))))))))))))).)).....))))......... ( -32.90) >DroEre_CAF1 5939 103 + 1 AUAAUGGCUGAUCCUGCGGCAGGAGCUCCAUCG---CCUCCAGCAGC---UGCUGCAAUGGCUCCUGUGGCAGCAAUAGCAGCAAUUCCGCCACCAGCUGAGCAGCUCU ....((((.((..((((.((.(((((......)---.)))).)).((---(((..((........))..)))))....))))....)).))))..((((....)))).. ( -37.80) >DroYak_CAF1 11639 103 + 1 AUUAUGGCCGAUCCUGCGGCAGGCGCUCCAUCG---CCCCAGGCACC---UGCUGCAAUGGCUCCAGUGGCACCAAUAGCAGCAAUUCCGCCACCAGCUGAGCAGCUCU .....(((((....(((((((((.(((......---.....))).))---))))))).)))))...(((((..................))))).((((....)))).. ( -34.17) >DroAna_CAF1 5677 86 + 1 AUUCUGGCCAGUCCGGCAGGAGGAGCACCAGGAGGAGCACCAGC------------AGUAGCACCAGUAGCUCCAGUAGCAUC------GCCCCUGGU-----GGCUCC ..(((.(((.....))).)))(((((((((((.(((((((..((------------....))....)).)))))....((...------)).))))))-----.))))) ( -34.60) >consensus AUAAUGGCAGAUCCUGCGGCAGGAGCUCCAUCG___CCUCCAGCACC___UGCUGCAAUGGCUCCUGUGGCACCAGUAGCAGCAAUUCCGCCACCAGCAGAGCAGCUCU ...((((....(((((...)))))...))))....................((((...(((((...(((((..................))))).)))))..))))... (-15.88 = -17.29 + 1.41)


| Location | 12,432,504 – 12,432,610 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 72.53 |
| Mean single sequence MFE | -43.16 |
| Consensus MFE | -20.66 |
| Energy contribution | -22.60 |
| Covariance contribution | 1.94 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.48 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.912951 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
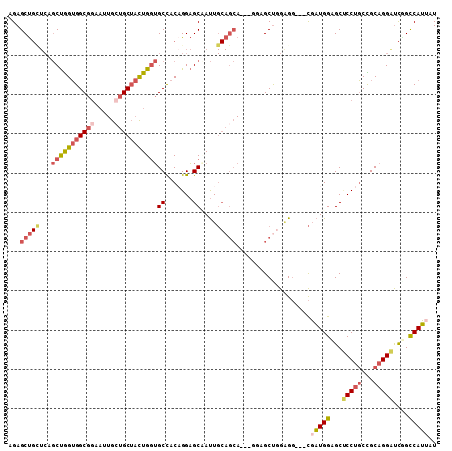
>X_DroMel_CAF1 12432504 106 - 22224390 AGAGCUGAUCUGCUGGUGGCGGAAUUGGUGCUACUGGUGCCACAGGAGCAAUUGCAGCAGCAGGAGCUGGAGG---CGAUGGAGCUCCUGCUGCAGGAUCUGCCAUUAU ..(((......)))(((((((((.....((((.(((......))).))))......((((((((((((.....---......))))))))))))....))))))))).. ( -48.40) >DroSec_CAF1 5889 106 - 1 AGAGCUGUUCUGCUGGUGGCGGAAUUGCUGCGACUGGUGCCACAGGAGCAAUUGCAGCAACAGGAGCUGGAGG---CGAUGGAGCCCCUGCCGCAGGGUCUGUCAUUAU ..(((......)))(((((((((....(((((.(((.(((..(((......)))..))).)))..((.((.((---(......))))).)))))))..))))))))).. ( -39.10) >DroEre_CAF1 5939 103 - 1 AGAGCUGCUCAGCUGGUGGCGGAAUUGCUGCUAUUGCUGCCACAGGAGCCAUUGCAGCA---GCUGCUGGAGG---CGAUGGAGCUCCUGCCGCAGGAUCAGCCAUUAU .(.((((.((....((((((((.....)))))))).((((..((((((((((((((((.---...)))....)---)))))..)))))))..)))))).)))))..... ( -42.70) >DroYak_CAF1 11639 103 - 1 AGAGCUGCUCAGCUGGUGGCGGAAUUGCUGCUAUUGGUGCCACUGGAGCCAUUGCAGCA---GGUGCCUGGGG---CGAUGGAGCGCCUGCCGCAGGAUCGGCCAUAAU ..((((....)))).(((((.((.(((((((...((((.((...)).))))..))))))---)...((((.((---((..((....)))))).)))).)).)))))... ( -45.80) >DroAna_CAF1 5677 86 - 1 GGAGCC-----ACCAGGGGC------GAUGCUACUGGAGCUACUGGUGCUACU------------GCUGGUGCUCCUCCUGGUGCUCCUCCUGCCGGACUGGCCAGAAU (((((.-----(((((((((------...)))...(((((.((..((......------------))..))))))).)))))))))))..(((((.....)).)))... ( -39.80) >consensus AGAGCUGCUCAGCUGGUGGCGGAAUUGCUGCUACUGGUGCCACAGGAGCAAUUGCAGCA___GGAGCUGGAGG___CGAUGGAGCUCCUGCCGCAGGAUCGGCCAUUAU ...(((((...(((((((((((.....)))))))))))((.......))....)))))...................(((((...(((((...)))))....))))).. (-20.66 = -22.60 + 1.94)


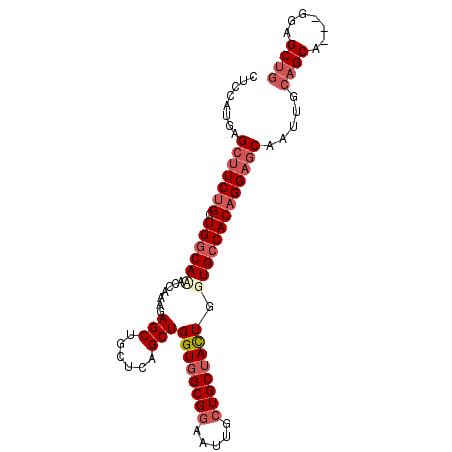
| Location | 12,432,541 – 12,432,637 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 86.74 |
| Mean single sequence MFE | -36.98 |
| Consensus MFE | -25.19 |
| Energy contribution | -27.38 |
| Covariance contribution | 2.19 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.681968 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12432541 96 - 22224390 CUCCAUGAGCUUCUGCUGGCAUACCAAAGAGCUGAUCUGCUGGUGGCGGAAUUGGUGCUACUGGUGCCACAGGAGCAAUUGCAGCAGCAGGAGCUG ((((....(((((((.((((..((((.((.((..((((((.....)))).))..)).))..)))))))))))))))...(((....)))))))... ( -39.40) >DroSec_CAF1 5926 96 - 1 CUCCAUGAGCUUCUGGUGACACACCAAAGAGCUGUUCUGCUGGUGGCGGAAUUGCUGCGACUGGUGCCACAGGAGCAAUUGCAGCAACAGGAGCUG ((((.(((((((.(((((...)))))..)))))(((((((.....)))))))((((((((.((.(.(....).).)).)))))))).))))))... ( -36.60) >DroEre_CAF1 5976 93 - 1 CUCCUUGAGGUUCUGCUGGCAAACCAAAGAGCUGCUCAGCUGGUGGCGGAAUUGCUGCUAUUGCUGCCACAGGAGCCAUUGCAGCA---GCUGCUG ........((((((..(((((........((((....))))((((((((.....))))))))..)))))..))))))....((((.---...)))) ( -32.90) >DroYak_CAF1 11676 93 - 1 CUCCAUGAGCCUCUGCUGGCAGACCAAAGAGCUGCUCAGCUGGUGGCGGAAUUGCUGCUAUUGGUGCCACUGGAGCCAUUGCAGCA---GGUGCCU ........(((.((((((((((.(......))))).)))).)).)))((..(((((((...((((.((...)).))))..))))))---)...)). ( -39.00) >consensus CUCCAUGAGCUUCUGCUGGCAAACCAAAGAGCUGCUCAGCUGGUGGCGGAAUUGCUGCUACUGGUGCCACAGGAGCAAUUGCAGCA___GGAGCUG ........(((((((.((((((.......(((......)))((((((((.....)))))))).))))))))))))).....((((.......)))) (-25.19 = -27.38 + 2.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:33:06 2006