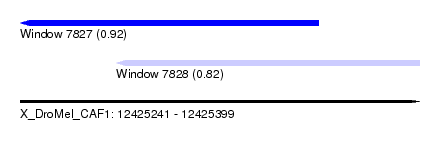
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,425,241 – 12,425,399 |
| Length | 158 |
| Max. P | 0.918810 |

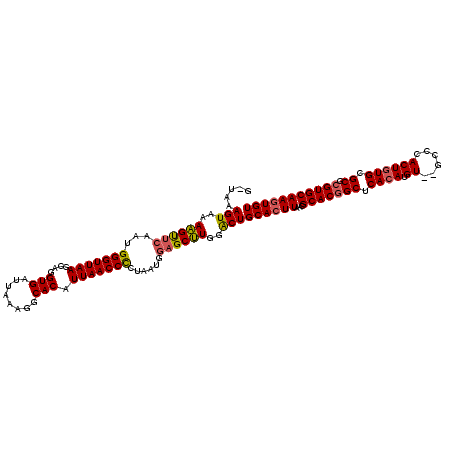
| Location | 12,425,241 – 12,425,359 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.31 |
| Mean single sequence MFE | -40.43 |
| Consensus MFE | -33.10 |
| Energy contribution | -33.67 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918810 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12425241 118 - 22224390 GUUAAAGUAAAAGUUCAAUGGGUUAAGCAGGUGAUUAAAGGCACAUUAACCCCUAAUGGAGCUUGGACUGCACUUAACGCACGGCUCACAUGU--GACCACUGUGCGCGCGUGCAAGUGU .....(((..((((((...(((((((....(((........))).)))))))......))))))..)))((((((...(((((((.((((.((--....)))))).)).))))))))))) ( -41.20) >DroSim_CAF1 4108 120 - 1 GCUAAAGUAAAAGUUCAAGGGGUUAAGCAGGUGAUUAAAGGCACAUUAACCCCUAAUGGAGCUUGGACUGCACUUAACGCACGGCUCACAUGUGUGCCCACUGUGCGCGCGUGCAAGUGU .....(((..((((((.(((((((((....(((........))).)))))))))....))))))..)))((((((...(((((((.((((.(((....))))))).)).))))))))))) ( -48.40) >DroYak_CAF1 4253 116 - 1 --UAAAGUAAAUGCCUAAUGGGUUAAGCAGGUGAUCAAAAGCACAUUAACCUCCAUUGUUGCAUAGGCUGCAAUUAGCGCACCGCUCAAAUGU--GCCCACUGUGCGCGCGUGCAAGUGU --...(((..((((.(((((((((((....(((........))).)))))..))))))..))))..)))(((....(((((((((......))--)......))))))...)))...... ( -31.70) >consensus G_UAAAGUAAAAGUUCAAUGGGUUAAGCAGGUGAUUAAAGGCACAUUAACCCCUAAUGGAGCUUGGACUGCACUUAACGCACGGCUCACAUGU__GCCCACUGUGCGCGCGUGCAAGUGU .....(((..((((((...(((((((....(((........))).)))))))......))))))..)))((((((...(((((((.((((.((......)))))).)).))))))))))) (-33.10 = -33.67 + 0.56)

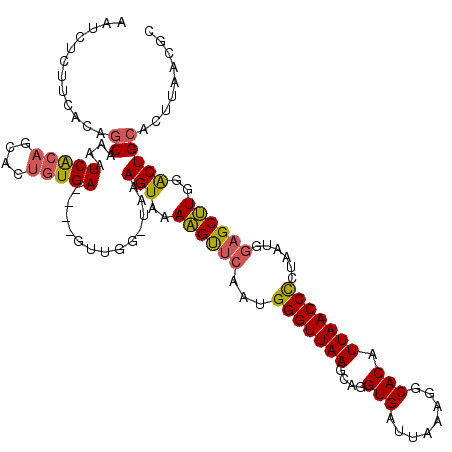
| Location | 12,425,279 – 12,425,399 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.84 |
| Mean single sequence MFE | -28.67 |
| Consensus MFE | -20.09 |
| Energy contribution | -20.43 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.815491 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12425279 120 - 22224390 AAUCUCUUCAAAGCAAAAUCACAGCACUGUGAUAUGGUUGGUUAAAGUAAAAGUUCAAUGGGUUAAGCAGGUGAUUAAAGGCACAUUAACCCCUAAUGGAGCUUGGACUGCACUUAACGC ............((...((((((....))))))...(((((....(((..((((((...(((((((....(((........))).)))))))......))))))..)))....))))))) ( -30.10) >DroSim_CAF1 4148 116 - 1 AUUCUCUUCACAUCAAAAUCACAGCACUGUGA----GUUGGCUAAAGUAAAAGUUCAAGGGGUUAAGCAGGUGAUUAAAGGCACAUUAACCCCUAAUGGAGCUUGGACUGCACUUAACGC ..................(((((....)))))----(((((....(((..((((((.(((((((((....(((........))).)))))))))....))))))..)))....))))).. ( -33.60) >DroYak_CAF1 4291 105 - 1 AGUUUUCUUACUGCAAAUUCG-----CUGUGA----------UAAAGUAAAUGCCUAAUGGGUUAAGCAGGUGAUCAAAAGCACAUUAACCUCCAUUGUUGCAUAGGCUGCAAUUAGCGC ...................((-----(((...----------...(((..((((.(((((((((((....(((........))).)))))..))))))..))))..))).....))))). ( -22.30) >consensus AAUCUCUUCACAGCAAAAUCACAGCACUGUGA____GUUGG_UAAAGUAAAAGUUCAAUGGGUUAAGCAGGUGAUUAAAGGCACAUUAACCCCUAAUGGAGCUUGGACUGCACUUAACGC ............((....(((((....))))).............(((..((((((...(((((((....(((........))).)))))))......))))))..)))))......... (-20.09 = -20.43 + 0.34)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:33:02 2006