
| Sequence ID | X_DroMel_CAF1 |
|---|---|
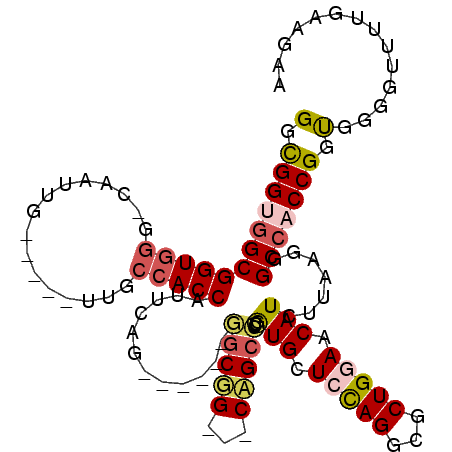
| Location | 12,420,800 – 12,420,898 |
| Length | 98 |
| Max. P | 0.870357 |

| Location | 12,420,800 – 12,420,898 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 81.04 |
| Mean single sequence MFE | -42.95 |
| Consensus MFE | -26.40 |
| Energy contribution | -26.81 |
| Covariance contribution | 0.41 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.533715 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
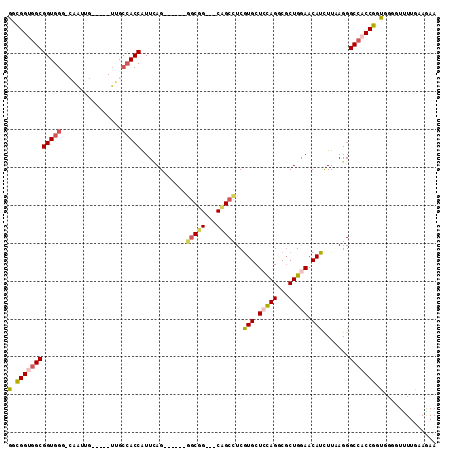
>X_DroMel_CAF1 12420800 98 + 22224390 GGUGGUGGCGGUGGG-CAAUUG-----UUGCCACCAUUCAG------GGCUG---CAGCCUCGUGCUCCAGGCGCUGAAACAUCUUAAGGGCCACCGGUGGGGUUUUGAAAAA (((((..(((((...-..))))-----)..))))).(((((------((((.---((.((..((((.....)))).............((....)))))).)))))))))... ( -39.00) >DroSec_CAF1 14849 98 + 1 GGUGGUGGCGGUGGG-CAAUUG-----UUGCCACCAUUCAG------GGCUG---CAGCCUCGUGCUCCAGGCGCUGAAACAUCUUAAGGGCCACCGGUGGGGUUUUGAAGAA (((((..(((((...-..))))-----)..))))).(((((------(.(((---.(((.....))).))).).)))))...((((.((((((.((...)))))))).)))). ( -40.40) >DroEre_CAF1 15150 98 + 1 GGCGGUGGCGGUGGG-CAAUUG-----UUGCCACCAUUCAG------AACUG---CAGCUUCGUGCUCCAGACGCUGGAACAUCUUAAGGGCCACCGGUGGGGUUUUGAAGAA ...((((((((..(.-...)..-----)))))))).(((((------((((.---((.(...(((.(((((...))))).))).....((....))).)).)))))))))... ( -38.60) >DroYak_CAF1 14806 98 + 1 GGUGGUGGCGGUGGG-CAAUUG-----UUGCCACCAUUUAG------GGCGG---CAGCCUCGUGCUCUAGGCGCUGGAACAUUUUCAGGGCCACCGGUGGCGUUUUGAAGAA (((((..(((((...-..))))-----)..))))).(((((------((((.---((.((..(....)..(((.(((((.....))))).)))...)))).)))))))))... ( -40.60) >DroAna_CAF1 15500 107 + 1 GGCGGCAGCGGUGGUGGUGUUGUGGUCUGGCCACCAUUCAG------AGCGGCGGCCGCCUCGUGCUCCAGCCGCUGGAACAUCUUCAGGGCCACCGGCGGGGUCUUGAAGAA ((((((.((.((..(((....((((.....))))...))).------.)).)).))))))...((.(((((...))))).))(((((((((((.((...))))))))))))). ( -51.10) >DroPer_CAF1 18600 97 + 1 GGCGGCGGCGG-GGA-CAGUUG-----------CCGUUGAGGCCGGCGGCGG---CCGCCUCAUGCUCCAGCCUCUGGAACAUCUUCAGGGCCACCGGCGGUGUCUUGAAAAA ((((((((((.-...-).))))-----------)))))((.((((.(((.((---((.....(((.(((((...))))).)))......)))).))).)))).))........ ( -48.00) >consensus GGCGGUGGCGGUGGG_CAAUUG_____UUGCCACCAUUCAG______GGCGG___CAGCCUCGUGCUCCAGGCGCUGGAACAUCUUAAGGGCCACCGGUGGGGUUUUGAAGAA (.((((((((((((................)))))............(((((...)))))..(((.(((((...))))).))).......))))))).).............. (-26.40 = -26.81 + 0.41)

| Location | 12,420,800 – 12,420,898 |
|---|---|
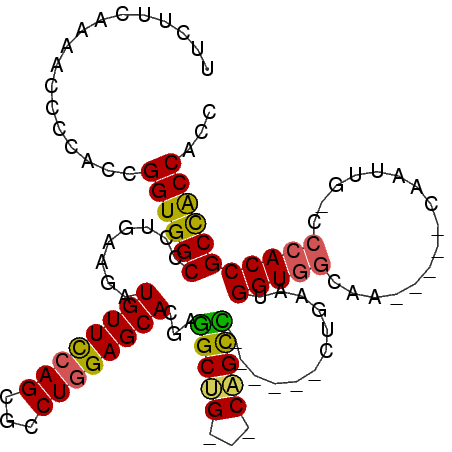
| Length | 98 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 81.04 |
| Mean single sequence MFE | -36.08 |
| Consensus MFE | -29.91 |
| Energy contribution | -29.21 |
| Covariance contribution | -0.71 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.870357 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12420800 98 - 22224390 UUUUUCAAAACCCCACCGGUGGCCCUUAAGAUGUUUCAGCGCCUGGAGCACGAGGCUG---CAGCC------CUGAAUGGUGGCAA-----CAAUUG-CCCACCGCCACCACC .................((((((...........(((((.(.(((.(((.....))).---))).)------))))).(((((((.-----....))-.)))))))))))... ( -35.20) >DroSec_CAF1 14849 98 - 1 UUCUUCAAAACCCCACCGGUGGCCCUUAAGAUGUUUCAGCGCCUGGAGCACGAGGCUG---CAGCC------CUGAAUGGUGGCAA-----CAAUUG-CCCACCGCCACCACC .................((((((...........(((((.(.(((.(((.....))).---))).)------))))).(((((((.-----....))-.)))))))))))... ( -35.20) >DroEre_CAF1 15150 98 - 1 UUCUUCAAAACCCCACCGGUGGCCCUUAAGAUGUUCCAGCGUCUGGAGCACGAAGCUG---CAGUU------CUGAAUGGUGGCAA-----CAAUUG-CCCACCGCCACCGCC ................(((((((.............(((...(((.(((.....))).---)))..------)))...(((((((.-----....))-.)))))))))))).. ( -33.50) >DroYak_CAF1 14806 98 - 1 UUCUUCAAAACGCCACCGGUGGCCCUGAAAAUGUUCCAGCGCCUAGAGCACGAGGCUG---CCGCC------CUAAAUGGUGGCAA-----CAAUUG-CCCACCGCCACCACC .................((((((..((....(((((.((...)).)))))...(((((---(((((------......))))))).-----.....)-))))..))))))... ( -30.70) >DroAna_CAF1 15500 107 - 1 UUCUUCAAGACCCCGCCGGUGGCCCUGAAGAUGUUCCAGCGGCUGGAGCACGAGGCGGCCGCCGCU------CUGAAUGGUGGCCAGACCACAACACCACCACCGCUGCCGCC .((((((.(.((((...)).)).).)))))).((..((((((.(((.((.....))((((((((..------.....))))))))..............)))))))))..)). ( -40.50) >DroPer_CAF1 18600 97 - 1 UUUUUCAAGACACCGCCGGUGGCCCUGAAGAUGUUCCAGAGGCUGGAGCAUGAGGCGG---CCGCCGCCGGCCUCAACGG-----------CAACUG-UCC-CCGCCGCCGCC ........((..(((.((((((((((....(((((((((...)))))))))..)).))---)))))).)))..))..(((-----------(.....-...-..))))..... ( -41.40) >consensus UUCUUCAAAACCCCACCGGUGGCCCUGAAGAUGUUCCAGCGCCUGGAGCACGAGGCUG___CAGCC______CUGAAUGGUGGCAA_____CAAUUG_CCCACCGCCACCACC .................((((((........((((((((...))))))))...(((((...)))))............(((((................)))))))))))... (-29.91 = -29.21 + -0.71)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:32:58 2006