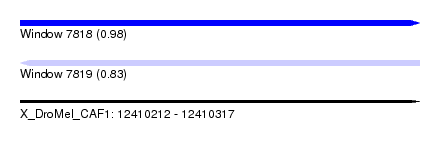
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,410,212 – 12,410,317 |
| Length | 105 |
| Max. P | 0.978051 |

| Location | 12,410,212 – 12,410,317 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 84.99 |
| Mean single sequence MFE | -21.24 |
| Consensus MFE | -15.90 |
| Energy contribution | -15.74 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.978051 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12410212 105 + 22224390 GCACUAGUGCACUUUCCC-AAAGGCAUUCUAAUUCUAAUUCCAACGAACCACUGUAUUCAGUUUCAGUUUACUGCAGUCGAACUUCACUUGACUGCAACUUCGUUG .....(((((.(((....-.)))))))).............(((((((..((((....))))..........(((((((((.......)))))))))..))))))) ( -25.40) >DroSec_CAF1 4359 106 + 1 ACACUAGUGCAUUUUUCCAAAAGGCAUUCUAAUUCUAAUUCCAACGAACCACUGUAUUCAGUUUCAGUUUACUGCAGUCGAACUUCACUUCACUGCAACUUCGUUG .....(((((.((((....))))))))).............(((((((..((((....))))..........((((((.(((......)))))))))..))))))) ( -20.30) >DroSim_CAF1 4405 94 + 1 ACACUAGUGCACUUUU------------CUAAUUCUAAUUCCAACGAACCACUGUAUUCAGUUUCAGUUUGCUGCAGUCGAACUUCACUUCACUGCAACUUCGUUG ................------------.............(((((((..((((....))))..........((((((.(((......)))))))))..))))))) ( -17.60) >DroEre_CAF1 4375 99 + 1 GUACUAAUACUAUUUUCA-AAUAGCAU------UCUAAUUCCAACGAACCACUGUAUUCAGUCUCAGUUUACUGCAGUCGAACUUCACUCGACUGCAACUUCGUUG .........(((((....-)))))...------........(((((((..((((....))))..........(((((((((.......)))))))))..))))))) ( -24.00) >DroYak_CAF1 4317 99 + 1 GCACUAGUGCUAUUUCCA-AAAUGCAU------UCCAAUUCCAACGAACCACUGUAUUCAGUUUCAGUUUUCUGCAGUCGAACUUCACUUAACUGCAACUUCGUUG (((....)))........-........------........(((((((..((((....))))..........((((((..(.......)..))))))..))))))) ( -18.90) >consensus GCACUAGUGCAAUUUUCA_AAAGGCAU_CUAAUUCUAAUUCCAACGAACCACUGUAUUCAGUUUCAGUUUACUGCAGUCGAACUUCACUUCACUGCAACUUCGUUG .........................................(((((((..((((....))))..........((((((.((.......)).))))))..))))))) (-15.90 = -15.74 + -0.16)


| Location | 12,410,212 – 12,410,317 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 84.99 |
| Mean single sequence MFE | -25.90 |
| Consensus MFE | -19.38 |
| Energy contribution | -19.14 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.834096 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12410212 105 - 22224390 CAACGAAGUUGCAGUCAAGUGAAGUUCGACUGCAGUAAACUGAAACUGAAUACAGUGGUUCGUUGGAAUUAGAAUUAGAAUGCCUUU-GGGAAAGUGCACUAGUGC (((((((.((((((((...........)))))))).........((((....))))..)))))))........(((((..((((((.-....))).)))))))).. ( -25.90) >DroSec_CAF1 4359 106 - 1 CAACGAAGUUGCAGUGAAGUGAAGUUCGACUGCAGUAAACUGAAACUGAAUACAGUGGUUCGUUGGAAUUAGAAUUAGAAUGCCUUUUGGAAAAAUGCACUAGUGU (((((((.((((((((((......))).))))))).........((((....))))..)))))))........(((((..(((..(((....))).)))))))).. ( -24.80) >DroSim_CAF1 4405 94 - 1 CAACGAAGUUGCAGUGAAGUGAAGUUCGACUGCAGCAAACUGAAACUGAAUACAGUGGUUCGUUGGAAUUAGAAUUAG------------AAAAGUGCACUAGUGU ((((((((((((((((((......))).))))))))........((((....))))..)))))))........(((((------------.........))))).. ( -24.40) >DroEre_CAF1 4375 99 - 1 CAACGAAGUUGCAGUCGAGUGAAGUUCGACUGCAGUAAACUGAGACUGAAUACAGUGGUUCGUUGGAAUUAGA------AUGCUAUU-UGAAAAUAGUAUUAGUAC (((((((.(((((((((((.....))))))))))).........((((....))))..))))))).......(------((((((((-....)))))))))..... ( -32.20) >DroYak_CAF1 4317 99 - 1 CAACGAAGUUGCAGUUAAGUGAAGUUCGACUGCAGAAAACUGAAACUGAAUACAGUGGUUCGUUGGAAUUGGA------AUGCAUUU-UGGAAAUAGCACUAGUGC (((((((.(((((((..((.....))..))))))).........((((....))))..)))))))..(((((.------.((((((.-....))).)))))))).. ( -22.20) >consensus CAACGAAGUUGCAGUCAAGUGAAGUUCGACUGCAGUAAACUGAAACUGAAUACAGUGGUUCGUUGGAAUUAGAAUUAG_AUGCCUUU_GGAAAAGUGCACUAGUGC (((((((.((((((((...........)))))))).........((((....))))..)))))))...............................(((....))) (-19.38 = -19.14 + -0.24)

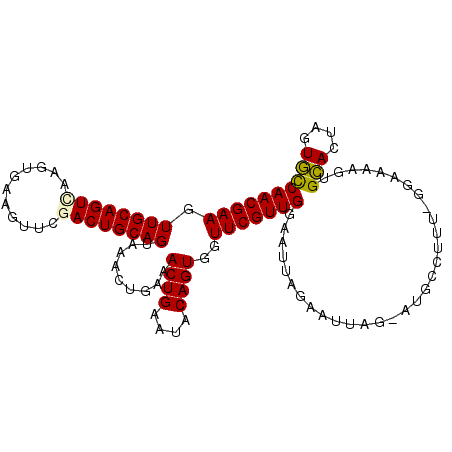

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:32:54 2006