
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,408,459 – 12,408,598 |
| Length | 139 |
| Max. P | 0.933018 |

| Location | 12,408,459 – 12,408,563 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 85.59 |
| Mean single sequence MFE | -27.11 |
| Consensus MFE | -19.15 |
| Energy contribution | -18.26 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.11 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933018 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12408459 104 + 22224390 GUAUGUGUAAUUUUAGAACUUGGCACGAAACAUGUUUCUAUGUAUAUUUUGUGUUGUGCACAU----AUAAACACGUAGCAUGUACUUGUACAUAUGUAUAUG------UACAU (((((((((............(((((((((((((....)))))....)))))))).)))))))----))...........((((((.(((((....))))).)------))))) ( -24.92) >DroEre_CAF1 2535 104 + 1 AUAUGUGUAAUUUUAGAACUUGGCACGAAACGUGUUUCUAUGUAUAUUUCGUGUUGUGCACAUAUAUAUAAACACGUAGCAUGUACUUCUACAUAUGUACAUAA---------- ...((((((....(((((....(((.(..(((((((..(((((((((...(((.....)))))))))))))))))))..).)))..)))))......)))))).---------- ( -26.00) >DroYak_CAF1 2482 110 + 1 GUAUGUGUAAUUUUAGAACUUGGCACGAAACAUGUUUCUAUGUAUAUUUUGUGUUGUGUACAU----AUAAACACGUAGAAUGUACUUGUACAUAUGUACAUACAUACAUACAA ((((((((.............(((((((((((((....)))))....))))))))((((((((----((..(((.(((.....))).)))..))))))))))..)))))))).. ( -30.40) >consensus GUAUGUGUAAUUUUAGAACUUGGCACGAAACAUGUUUCUAUGUAUAUUUUGUGUUGUGCACAU____AUAAACACGUAGCAUGUACUUGUACAUAUGUACAUA______UACA_ .(((((((....(((.....((((((((.(((.................))).)))))).))......))))))))))..((((((..........))))))............ (-19.15 = -18.26 + -0.88)

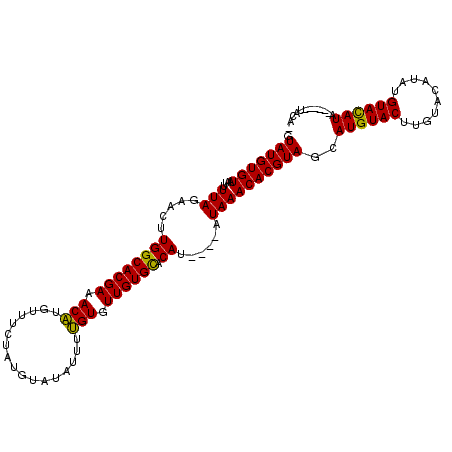

| Location | 12,408,496 – 12,408,598 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 83.49 |
| Mean single sequence MFE | -20.56 |
| Consensus MFE | -16.35 |
| Energy contribution | -15.69 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.633038 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12408496 102 + 22224390 CUAUGUAUAUUUUGUGUUGUGCACAU----AUAAACACGUAGCAUGUACUUGUACAUAUGUAUAUG------UACAUAUACCCACAUUCACUUAUUACUUGGCAACGCCGGC ((((((......((((.....)))).----......)))))).((((((.(((((....))))).)------))))).....................(((((...))))). ( -21.22) >DroEre_CAF1 2572 100 + 1 CUAUGUAUAUUUCGUGUUGUGCACAUAUAUAUAAACACGUAGCAUGUACUUCUACAUAUGUACAUAA------------AUUCACAUUCACUUAUUACUUGGCAACGCCGGC .(((((((((..((((((...............))))))....(((((....)))))))))))))).------------...................(((((...))))). ( -16.86) >DroYak_CAF1 2519 108 + 1 CUAUGUAUAUUUUGUGUUGUGUACAU----AUAAACACGUAGAAUGUACUUGUACAUAUGUACAUACAUACAUACAAAUAUCCACAUUCACUUAUUACUUGGCGACGCCGGC ..((((((((((((((((((((((((----((..(((.(((.....))).)))..)))))))))))...))))).))))))..))))...........(((((...))))). ( -23.60) >consensus CUAUGUAUAUUUUGUGUUGUGCACAU____AUAAACACGUAGCAUGUACUUGUACAUAUGUACAUA______UACA_AUAUCCACAUUCACUUAUUACUUGGCAACGCCGGC .(((((((((..((((((...............))))))....(((((....))))))))))))))................................(((((...))))). (-16.35 = -15.69 + -0.66)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:32:52 2006