| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,402,664 – 12,402,775 |
| Length | 111 |
| Max. P | 0.933805 |

| Location | 12,402,664 – 12,402,775 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 75.04 |
| Mean single sequence MFE | -28.31 |
| Consensus MFE | -15.16 |
| Energy contribution | -14.85 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933805 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12402664 111 + 22224390 UCGGUGGUAAAUGCAUAGAAAAUUGAGAAUGAAUUUACCAAAACGAUAAGCCAAGAGCUUUCUUUGUGGGCAAAUCGCAAAGAACACCACCA--AUAGCGGGCGCUGUGGUAU (((.((((((((.(((............))).))))))))...)))...((((..(((.((((((((((.....)))))))))).....((.--.....))..))).)))).. ( -31.30) >DroSec_CAF1 5347 111 + 1 UCGGUGGUAAAUGCAUAGAAAAUUGAGAAUGAAUUUACCAUAACAAUAUGCUGAAAGCUUCCAUUAUGGACAAAUAGCAAAGAACGCCACCA--AUAGCGGGCGCUGUGGUAU ..((((((((((.(((............))).))))))).........(((((......(((.....)))....)))))......)))((((--.((((....)))))))).. ( -25.80) >DroEre_CAF1 5780 102 + 1 UCGGUGGUAAAUGCA--AAAAAU-----UUAGAUU----AAAUAAAUAAGCUAAAAGCUUCUUUUAUGGACAAUUUGCUAAGAACUCCACCACCACAGCGGGUGCUGUGGUAU ..(((((.....(((--((..((-----(((....----...)))))((((.....)))).............)))))........)))))((((((((....)))))))).. ( -28.92) >DroYak_CAF1 5650 100 + 1 UCGGUGGUAAAUUGA--GA---------UUAGAUUCUCAAAAAAAAUAAGCUGGAAGCUUCCCCUAUGAACAAAUCGCUUGGAACGCCACCA--AUAGCGGGCGCUGUGGUAU ..((((((...((((--((---------......))))))......(((((.(((....))).....((.....)))))))....)))))).--(((((....)))))..... ( -27.20) >consensus UCGGUGGUAAAUGCA__GAAAAU_____AUAAAUUUACCAAAAAAAUAAGCUAAAAGCUUCCUUUAUGGACAAAUCGCAAAGAACGCCACCA__AUAGCGGGCGCUGUGGUAU ..((((((.........................................((.....)).(((.....)))...............))))))...(((((....)))))..... (-15.16 = -14.85 + -0.31)

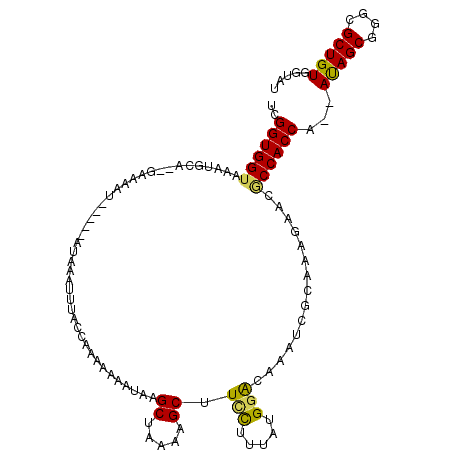
| Location | 12,402,664 – 12,402,775 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 75.04 |
| Mean single sequence MFE | -25.35 |
| Consensus MFE | -13.48 |
| Energy contribution | -13.85 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.544518 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12402664 111 - 22224390 AUACCACAGCGCCCGCUAU--UGGUGGUGUUCUUUGCGAUUUGCCCACAAAGAAAGCUCUUGGCUUAUCGUUUUGGUAAAUUCAUUCUCAAUUUUCUAUGCAUUUACCACCGA ((((((((((....)))..--..))))))).....(((((..(((....((((....)))))))..)))))..((((((((.(((............))).)))))))).... ( -28.60) >DroSec_CAF1 5347 111 - 1 AUACCACAGCGCCCGCUAU--UGGUGGCGUUCUUUGCUAUUUGUCCAUAAUGGAAGCUUUCAGCAUAUUGUUAUGGUAAAUUCAUUCUCAAUUUUCUAUGCAUUUACCACCGA .......((((((.((...--..))))))))...((((.....(((.....))).......))))........((((((((.(((............))).)))))))).... ( -22.50) >DroEre_CAF1 5780 102 - 1 AUACCACAGCACCCGCUGUGGUGGUGGAGUUCUUAGCAAAUUGUCCAUAAAAGAAGCUUUUAGCUUAUUUAUUU----AAUCUAA-----AUUUUU--UGCAUUUACCACCGA ..((((((((....))))))))(((((..(((((................)))))((.(((((.(((......)----)).))))-----).....--.)).....))))).. ( -26.89) >DroYak_CAF1 5650 100 - 1 AUACCACAGCGCCCGCUAU--UGGUGGCGUUCCAAGCGAUUUGUUCAUAGGGGAAGCUUCCAGCUUAUUUUUUUUGAGAAUCUAA---------UC--UCAAUUUACCACCGA .......(((....))).(--((((((......((((((.....)).....(((....))).)))).......((((((......---------))--))))....))))))) ( -23.40) >consensus AUACCACAGCGCCCGCUAU__UGGUGGCGUUCUUAGCGAUUUGUCCAUAAAGGAAGCUUUCAGCUUAUUGUUUUGGUAAAUCCAA_____AUUUUC__UGCAUUUACCACCGA .......(((....)))....((((((.((.....)).......(((.(((((((((.....)))).))))).)))..............................)))))). (-13.48 = -13.85 + 0.37)

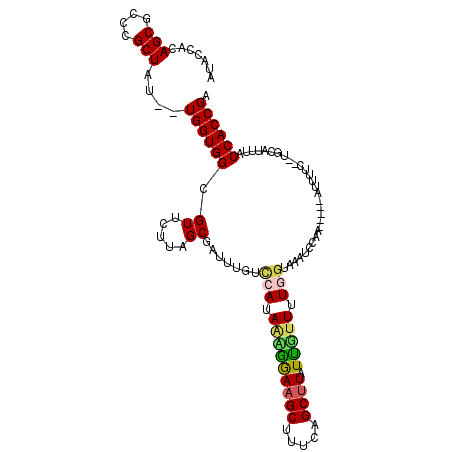
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:32:48 2006