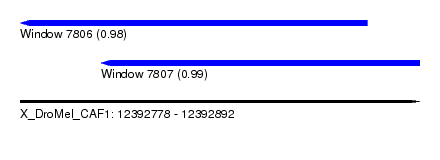
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,392,778 – 12,392,892 |
| Length | 114 |
| Max. P | 0.988276 |

| Location | 12,392,778 – 12,392,877 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 64.81 |
| Mean single sequence MFE | -26.40 |
| Consensus MFE | -12.62 |
| Energy contribution | -12.66 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.48 |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.981113 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
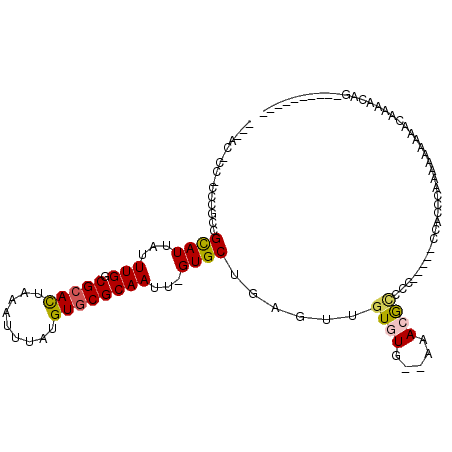
>X_DroMel_CAF1 12392778 99 - 22224390 ACGACGCCACCCGCCGCAUAAUUUGGCGCACUAAAUUUAUGUGCGCAAUU-GUGCUGAGUUGUGUGGGGAAGGCCCC-----CCCCCCAAAAAAAAAAACACCAG---------- .....(((..((.((((((((((..((((((.........))))((....-))))..))))))))))))..)))...-----.......................---------- ( -32.60) >DroGri_CAF1 5633 91 - 1 ----------CUUCCGUAUUAUUUGGCGCACUAAAUUUAUGUGCGCAAUU-GUGCUGAGUUGUGUG--AAACAUCCCCACAUGCACUCAUACACACAC-ACACAG---------- ----------...............((((((.........))))))...(-(((.((.((((((((--(..(((......)))...))))))).)).)-))))).---------- ( -23.20) >DroSec_CAF1 6529 100 - 1 ACGACGCCACCCGCCGCAUAAUUUGGCGCACUAAAUUUAUGUGCGCAAUUAGUGCUGAGUUGUGUGGGGAAGGCACC-----CCCCCCAAAAAAAAAAAAAANNN---------- .....(((..((.((((((((((..((((((.........))))((.....))))..))))))))))))..)))...-----.......................---------- ( -33.20) >DroMoj_CAF1 10045 82 - 1 ----------CUUGCGUAUUAUUUGGCGCAUUAAAUUUAUGUGCGCAAUU-GUGCUGAGUUGUGUG--AAACAGCC-CAC--CCACCCACACAGCCAC----------------- ----------.....((((...(((((((((.......)))))).)))..-))))((.((((((((--........-...--.....)))))))))).----------------- ( -21.99) >DroAna_CAF1 7431 92 - 1 ---ACUCCGCCCGCCGCAUUACUUGCCGCACUAAAUUUAUGUUCGCAAUU-GUGCUGAGUUGUGUG--CAACGCCCC-----------G------AACACACCAGCAGGAUAUUU ---..(((((..((.(((.....))).))..........((((((((...-.)))...((((....--)))).....-----------)------)))).....)).)))..... ( -21.00) >consensus ___AC_CC_CCCGCCGCAUUAUUUGGCGCACUAAAUUUAUGUGCGCAAUU_GUGCUGAGUUGUGUG__AAACGCCCC_____CCACCCAAAAAAAAACAAAACAG__________ ...............((((...(((.(((((.........))))))))...))))......((((.....))))......................................... (-12.62 = -12.66 + 0.04)


| Location | 12,392,801 – 12,392,892 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 74.36 |
| Mean single sequence MFE | -28.74 |
| Consensus MFE | -17.82 |
| Energy contribution | -17.52 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.62 |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.988276 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12392801 91 - 22224390 UGUGC--CAUUCACUU--------------CACGACGCCACCCGCCGCAUAAUUUGGCGCACUAAAUUUAUGUGCGCAAUU-GUGCUGAGUUGUGUGGGGAAGGCCCC--- ...((--(.(((.((.--------------((((((((.....)).(((((((...((((((.........)))))).)))-))))...)))))).)).))))))...--- ( -34.20) >DroGri_CAF1 5657 83 - 1 UAUGC--GUUUUACUUGU-----------------------CUUCCGUAUUAUUUGGCGCACUAAAUUUAUGUGCGCAAUU-GUGCUGAGUUGUGUG--AAACAUCCCCAC .....--(((((((....-----------------------.(((.((((......((((((.........))))))....-)))).)))....)))--))))........ ( -21.80) >DroSec_CAF1 6552 104 - 1 UGUGC--CAUUCACUUCACCUUUUC--CUUCACGACGCCACCCGCCGCAUAAUUUGGCGCACUAAAUUUAUGUGCGCAAUUAGUGCUGAGUUGUGUGGGGAAGGCACC--- .((((--(.............((((--((.((((((((.....)).((((......((((((.........)))))).....))))...)))))).))))))))))).--- ( -35.21) >DroEre_CAF1 6729 98 - 1 UGUGC--CAUUCACUCGACUUUUUU--GUUC---ACGCCACCCGCCGCAUAAUUUGGCGCACUAAAUUUAUGUGCGCAAUU-GUGCUGAGUUGUGCG--AAACGCCCC--- ...((--..(((.(.((((((....--....---..((.....)).(((((((...((((((.........)))))).)))-)))).)))))).).)--))..))...--- ( -26.90) >DroWil_CAF1 37534 80 - 1 UAUGC--GUUUCACUUG-----------------------CCUUGCGUAUUAUUUGGCGCAUUAAAUUUAUGUGCGCAAUU-GUGCUGAGUUGUGUG--UAACGUGUC--- .((((--(((.(((.(.-----------------------.((((.((((...(((((((((.......)))))).)))..-))))))))..).)))--.))))))).--- ( -22.50) >DroYak_CAF1 11349 102 - 1 UGUGCCCCAUUCACUUGACUUUUUUGUGUUC---ACGCCACCCGCCGCAUAAUUUGGCGCACUAAAUUUAUGUGCGCAAUU-GUGCCGAGUUGUGUG--AAACGCCCC--- .(((.......)))...........((((((---((((.((.((..(((((((...((((((.........)))))).)))-)))))).)).)))))--.)))))...--- ( -31.80) >consensus UGUGC__CAUUCACUUGA____________C___ACGCCACCCGCCGCAUAAUUUGGCGCACUAAAUUUAUGUGCGCAAUU_GUGCUGAGUUGUGUG__AAACGCCCC___ .............................................(((((((((((((((((.........))))((.....))))))))))))))).............. (-17.82 = -17.52 + -0.30)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:32:43 2006