
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,383,334 – 12,383,438 |
| Length | 104 |
| Max. P | 0.970814 |

| Location | 12,383,334 – 12,383,438 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 85.67 |
| Mean single sequence MFE | -34.63 |
| Consensus MFE | -30.89 |
| Energy contribution | -30.67 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970814 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
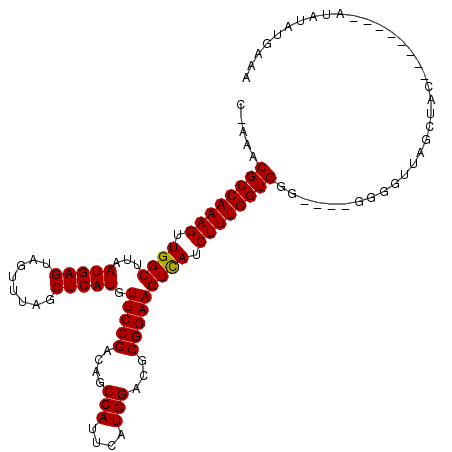
>X_DroMel_CAF1 12383334 104 + 22224390 CCAAACGCCAAAGUUGGCUUAAUGAGAAGUUUAGCUCAUGUUCCCACAGCCAUUCAUGGAUGGGGAAGUUAUCUUUGGUGGGGUGGGGGGUUGGCU------------AUAUGAAA (((..((((((((.((((...(((((........))))).(((((.((.(((....))).))))))))))).))))))))...)))..........------------........ ( -33.80) >DroSim_CAF1 15204 103 + 1 C-AAACGCCAAAGUUGGCUUAAUGAGUAGUUUAGCUCAUGUUCCCACAGCCAUUCAUGGACGGGGAAGUCAUCUUUGGUGGG----GGGGUUAGCUAC--------AUAUAUGAAA .-...((((((((.((((...((((((......)))))).(((((....(((....)))...))))))))).))))))))..----............--------.......... ( -32.70) >DroYak_CAF1 2465 110 + 1 G-AAACGCCAAAGUUGGCUUAAUGAGUAGUUUAGCUCAUGUUCCCACAGCCAUUCAUGGACGGGGAAGUCAACUUUGGUGG-----GGGGUUAGCUACAUACAUACAUAUAUGAAA .-...(((((((((((((...((((((......)))))).(((((....(((....)))...)))))))))))))))))).-----...............((((....))))... ( -37.40) >consensus C_AAACGCCAAAGUUGGCUUAAUGAGUAGUUUAGCUCAUGUUCCCACAGCCAUUCAUGGACGGGGAAGUCAUCUUUGGUGGG____GGGGUUAGCUAC________AUAUAUGAAA .....((((((((.((((...(((((........))))).(((((....(((....)))...))))))))).)))))))).................................... (-30.89 = -30.67 + -0.22)

| Location | 12,383,334 – 12,383,438 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 85.67 |
| Mean single sequence MFE | -23.93 |
| Consensus MFE | -20.69 |
| Energy contribution | -20.47 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.560720 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12383334 104 - 22224390 UUUCAUAU------------AGCCAACCCCCCACCCCACCAAAGAUAACUUCCCCAUCCAUGAAUGGCUGUGGGAACAUGAGCUAAACUUCUCAUUAAGCCAACUUUGGCGUUUGG ........------------..........(((...(.((((((.....(((((((.(((....))).)).))))).(((((........)))))........)))))).)..))) ( -22.50) >DroSim_CAF1 15204 103 - 1 UUUCAUAUAU--------GUAGCUAACCCC----CCCACCAAAGAUGACUUCCCCGUCCAUGAAUGGCUGUGGGAACAUGAGCUAAACUACUCAUUAAGCCAACUUUGGCGUUU-G ..........--------............----..(.((((((.((..(((((((.(((....))).)).))))).(((((........))))).....)).)))))).)...-. ( -21.40) >DroYak_CAF1 2465 110 - 1 UUUCAUAUAUGUAUGUAUGUAGCUAACCCC-----CCACCAAAGUUGACUUCCCCGUCCAUGAAUGGCUGUGGGAACAUGAGCUAAACUACUCAUUAAGCCAACUUUGGCGUUU-C ...((((((....))))))...........-----.(.(((((((((..(((((((.(((....))).)).))))).(((((........))))).....))))))))).)...-. ( -27.90) >consensus UUUCAUAUAU________GUAGCUAACCCC____CCCACCAAAGAUGACUUCCCCGUCCAUGAAUGGCUGUGGGAACAUGAGCUAAACUACUCAUUAAGCCAACUUUGGCGUUU_G ....................................(.((((((.....(((((((.(((....))).)).))))).(((((........)))))........)))))).)..... (-20.69 = -20.47 + -0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:32:36 2006