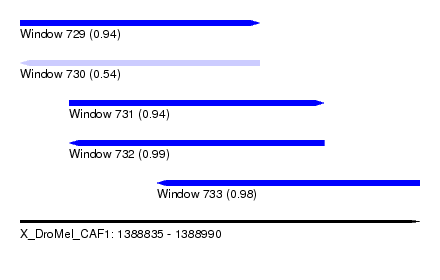
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,388,835 – 1,388,990 |
| Length | 155 |
| Max. P | 0.992285 |

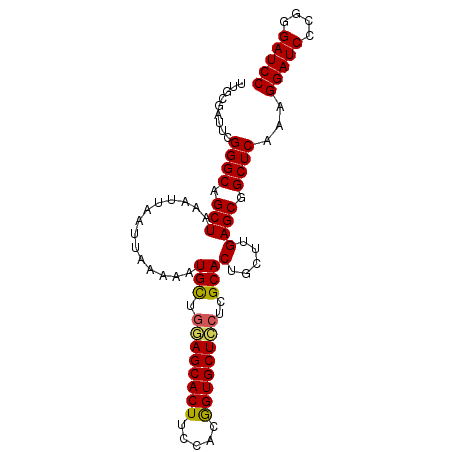
| Location | 1,388,835 – 1,388,928 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 87.81 |
| Mean single sequence MFE | -31.32 |
| Consensus MFE | -26.66 |
| Energy contribution | -26.48 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.940611 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
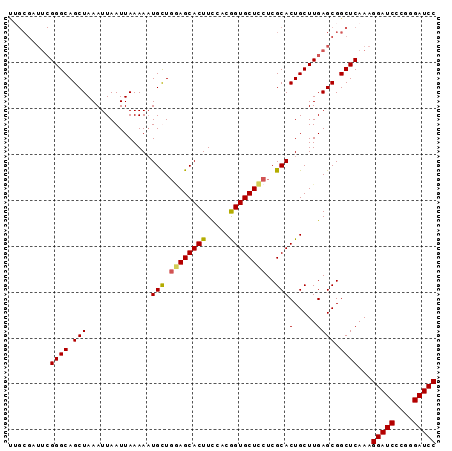
>X_DroMel_CAF1 1388835 93 + 22224390 UUGCGAUUCGGGCAGCUAAAUUAAUUAAAAAUGUUGGAGCACUUCUACGGUGCUCCUCGCACUGCUUGAGCGGCUCAAAGGAUCCCGGGAUCC ..((..(((((((((................(((.((((((((.....))))))))..))))))))))))..)).....(((((....))))) ( -32.69) >DroSec_CAF1 16156 93 + 1 UUGCGAUUCGGGCAGCUAAAUUAAUUAAAAAUGUUGGAGCACUUCCACGGUGCUCCUCGCACUGCUUGAGCGGCUCAAAGGAUCCCGGGAUCC ..((..(((((((((................(((.((((((((.....))))))))..))))))))))))..)).....(((((....))))) ( -32.69) >DroEre_CAF1 15088 93 + 1 CGGCGCUUCGGGCAGCUAAAUUAAUUAAAAUUGCUCAAGCACUUCCCCGGUGCUUCUCGCACUGCUUGAGCGGCUCAAAGGAUCCGGGGAUCC .(.((((.(((((((...((((......))))((..(((((((.....)))))))...)).))))))))))).).....(((((....))))) ( -32.60) >DroYak_CAF1 4413 84 + 1 ---------GGGCAGCUAAAUUAAUUAAAAAUGCUGCAGCACUUUCCCAGUGCUCCUCGCACUGCUUGAGCGGCUCAAAGGAUCCGGGGAUCC ---------..(((((................)))))(((.(((...((((((.....))))))...)))..)))....(((((....))))) ( -27.29) >consensus UUGCGAUUCGGGCAGCUAAAUUAAUUAAAAAUGCUGGAGCACUUCCACGGUGCUCCUCGCACUGCUUGAGCGGCUCAAAGGAUCCCGGGAUCC .........((((.(((..............(((.((((((((.....))))))))..)))(.....)))).))))...(((((....))))) (-26.66 = -26.48 + -0.19)

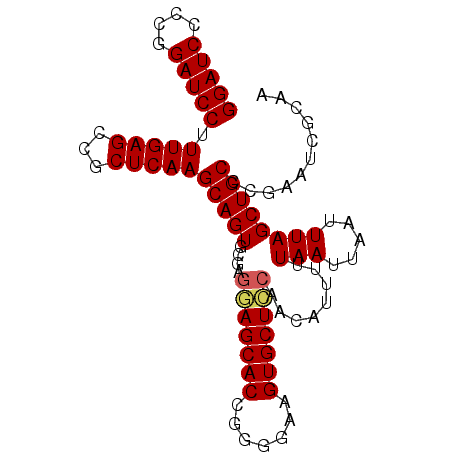
| Location | 1,388,835 – 1,388,928 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 87.81 |
| Mean single sequence MFE | -29.38 |
| Consensus MFE | -24.40 |
| Energy contribution | -24.65 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.540178 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1388835 93 - 22224390 GGAUCCCGGGAUCCUUUGAGCCGCUCAAGCAGUGCGAGGAGCACCGUAGAAGUGCUCCAACAUUUUUAAUUAAUUUAGCUGCCCGAAUCGCAA (((((....)))))((((((...))))))...((((((((((((.......))))))).......((((.....)))).........))))). ( -28.00) >DroSec_CAF1 16156 93 - 1 GGAUCCCGGGAUCCUUUGAGCCGCUCAAGCAGUGCGAGGAGCACCGUGGAAGUGCUCCAACAUUUUUAAUUAAUUUAGCUGCCCGAAUCGCAA (((((....)))))((((((...))))))...((((((((((((.......))))))).......((((.....)))).........))))). ( -28.00) >DroEre_CAF1 15088 93 - 1 GGAUCCCCGGAUCCUUUGAGCCGCUCAAGCAGUGCGAGAAGCACCGGGGAAGUGCUUGAGCAAUUUUAAUUAAUUUAGCUGCCCGAAGCGCCG (((((....)))))...(.((.(((((((((((((.....))))........)))))))))................(((......)))))). ( -31.80) >DroYak_CAF1 4413 84 - 1 GGAUCCCCGGAUCCUUUGAGCCGCUCAAGCAGUGCGAGGAGCACUGGGAAAGUGCUGCAGCAUUUUUAAUUAAUUUAGCUGCCC--------- (((((....))))).....((.(((.((.((((((.....)))))).((((((((....))))))))......)).))).))..--------- ( -29.70) >consensus GGAUCCCCGGAUCCUUUGAGCCGCUCAAGCAGUGCGAGGAGCACCGGGGAAGUGCUCCAACAUUUUUAAUUAAUUUAGCUGCCCGAAUCGCAA (((((....))))).(((((...)))))(((((....(((((((.......)))))))........(((.....))))))))........... (-24.40 = -24.65 + 0.25)

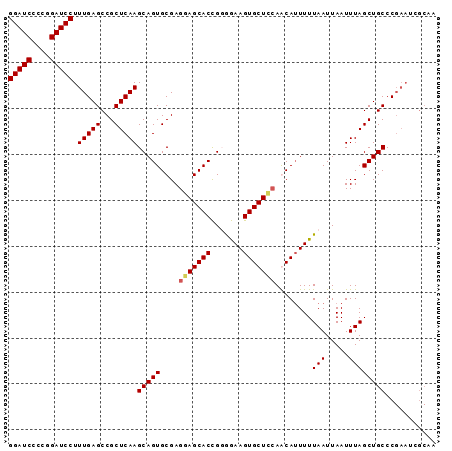
| Location | 1,388,854 – 1,388,953 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 85.90 |
| Mean single sequence MFE | -37.90 |
| Consensus MFE | -33.19 |
| Energy contribution | -33.50 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940665 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1388854 99 + 22224390 AUUAAUUAAAAAUGUUGGAGCACUUCUACGGUGCUCCUCGCACUGCUUGAGCGGCUCAAAGGAUCCCGGGAUCCUGUGUGCCCG------UGUCCGUGCCCGUGU ................((((((((.....))))))))..((((.((....))(((.((.((((((....)))))).)).)))..------.....))))...... ( -37.30) >DroSec_CAF1 16175 93 + 1 AUUAAUUAAAAAUGUUGGAGCACUUCCACGGUGCUCCUCGCACUGCUUGAGCGGCUCAAAGGAUCCCGGGAUCCUGUGUGCCCG------------UGCCCGUGU ................((((((((.....))))))))..((((.((....))(((.((.((((((....)))))).)).))).)------------)))...... ( -37.30) >DroEre_CAF1 15107 105 + 1 AUUAAUUAAAAUUGCUCAAGCACUUCCCCGGUGCUUCUCGCACUGCUUGAGCGGCUCAAAGGAUCCGGGGAUCCUGUGUGCCAGUGCCCGUGCCUGUGCCCGUGU .............(((((((((........((((.....)))))))))))))(((.((.((((((....)))))).)).)))...((.((.((....)).)).)) ( -38.70) >DroYak_CAF1 4423 105 + 1 AUUAAUUAAAAAUGCUGCAGCACUUUCCCAGUGCUCCUCGCACUGCUUGAGCGGCUCAAAGGAUCCGGGGAUCCUGUGUGCCAGUGGCGGUGGCUGUGCCCGUGU .............(((((.((.(((...((((((.....))))))...))).(((.((.((((((....)))))).)).))).)).)))))(((...)))..... ( -38.30) >consensus AUUAAUUAAAAAUGCUGGAGCACUUCCACGGUGCUCCUCGCACUGCUUGAGCGGCUCAAAGGAUCCCGGGAUCCUGUGUGCCAG______UG_CUGUGCCCGUGU ................((((((((.....))))))))..((((.((....))(((.((.((((((....)))))).)).))).............))))...... (-33.19 = -33.50 + 0.31)

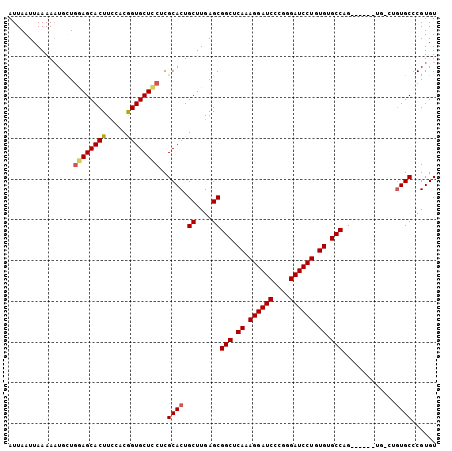
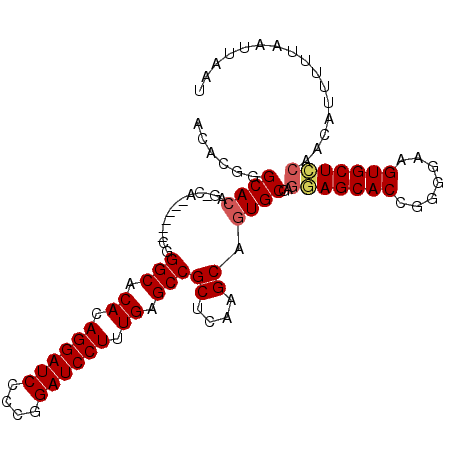
| Location | 1,388,854 – 1,388,953 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 85.90 |
| Mean single sequence MFE | -39.20 |
| Consensus MFE | -33.00 |
| Energy contribution | -33.50 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.32 |
| SVM RNA-class probability | 0.992285 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1388854 99 - 22224390 ACACGGGCACGGACA------CGGGCACACAGGAUCCCGGGAUCCUUUGAGCCGCUCAAGCAGUGCGAGGAGCACCGUAGAAGUGCUCCAACAUUUUUAAUUAAU ......((((.....------..(((.((.((((((....)))))).)).)))((....)).))))..(((((((.......)))))))................ ( -37.70) >DroSec_CAF1 16175 93 - 1 ACACGGGCA------------CGGGCACACAGGAUCCCGGGAUCCUUUGAGCCGCUCAAGCAGUGCGAGGAGCACCGUGGAAGUGCUCCAACAUUUUUAAUUAAU ......(((------------(.(((.((.((((((....)))))).)).)))((....)).))))..(((((((.......)))))))................ ( -37.70) >DroEre_CAF1 15107 105 - 1 ACACGGGCACAGGCACGGGCACUGGCACACAGGAUCCCCGGAUCCUUUGAGCCGCUCAAGCAGUGCGAGAAGCACCGGGGAAGUGCUUGAGCAAUUUUAAUUAAU ......((.(((((((((((...(((.((.((((((....)))))).)).)))))))...(.((((.....)))).).....))))))).))............. ( -41.00) >DroYak_CAF1 4423 105 - 1 ACACGGGCACAGCCACCGCCACUGGCACACAGGAUCCCCGGAUCCUUUGAGCCGCUCAAGCAGUGCGAGGAGCACUGGGAAAGUGCUGCAGCAUUUUUAAUUAAU ....((((...((....))....(((.((.((((((....)))))).)).)))))))...((((((.....)))))).((((((((....))))))))....... ( -40.40) >consensus ACACGGGCACAG_CA______CGGGCACACAGGAUCCCCGGAUCCUUUGAGCCGCUCAAGCAGUGCGAGGAGCACCGGGGAAGUGCUCCAACAUUUUUAAUUAAU ......((((.............(((.((.((((((....)))))).)).)))((....)).))))..(((((((.......)))))))................ (-33.00 = -33.50 + 0.50)

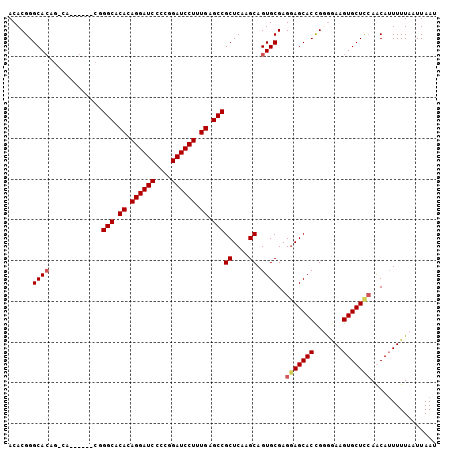
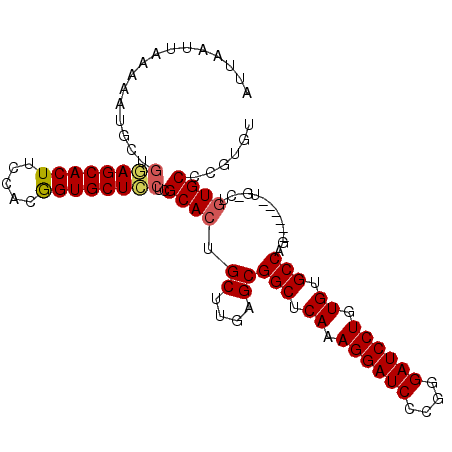
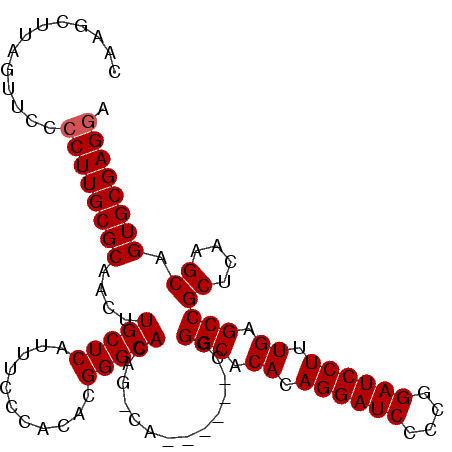
| Location | 1,388,888 – 1,388,990 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 89.72 |
| Mean single sequence MFE | -37.72 |
| Consensus MFE | -34.72 |
| Energy contribution | -34.98 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.979691 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1388888 102 - 22224390 CAAGCUUAGUUCCCCUUGCGCAACUUGCUCAUUUUCAACACGGGCACGGACA------CGGGCACACAGGAUCCCGGGAUCCUUUGAGCCGCUCAAGCAGUGCGAGGA .............((((((((....(((((...........)))))((....------))(((.((.((((((....)))))).)).)))((....)).)))))))). ( -37.00) >DroSec_CAF1 16209 96 - 1 CAAGCUUAGUUCCCCUUGCGCAACUUGCUCAUUUCCCACACGGGCA------------CGGGCACACAGGAUCCCGGGAUCCUUUGAGCCGCUCAAGCAGUGCGAGGA .............((((((((..((((..(....(((....)))..------------..(((.((.((((((....)))))).)).))))..))))..)))))))). ( -37.10) >DroEre_CAF1 15141 108 - 1 CAAGCUUAGUUCCCCUUGCGCAACUUGCUCAUUUCCCACACGGGCACAGGCACGGGCACUGGCACACAGGAUCCCCGGAUCCUUUGAGCCGCUCAAGCAGUGCGAGAA ..............(((((((....((((.....(((....)))....)))).((((...(((.((.((((((....)))))).)).))))))).....))))))).. ( -38.60) >DroYak_CAF1 4457 108 - 1 CAAGCUUAGUUCCCCUUGCGCAACUUGCUCAUUUCCCACACGGGCACAGCCACCGCCACUGGCACACAGGAUCCCCGGAUCCUUUGAGCCGCUCAAGCAGUGCGAGGA .............((((((((.....(((((...(((....)))....((((.......))))....((((((....)))))).))))).((....)).)))))))). ( -38.20) >consensus CAAGCUUAGUUCCCCUUGCGCAACUUGCUCAUUUCCCACACGGGCACAG_CA______CGGGCACACAGGAUCCCCGGAUCCUUUGAGCCGCUCAAGCAGUGCGAGGA .............((((((((....(((((...........)))))..............(((.((.((((((....)))))).)).)))((....)).)))))))). (-34.72 = -34.98 + 0.25)

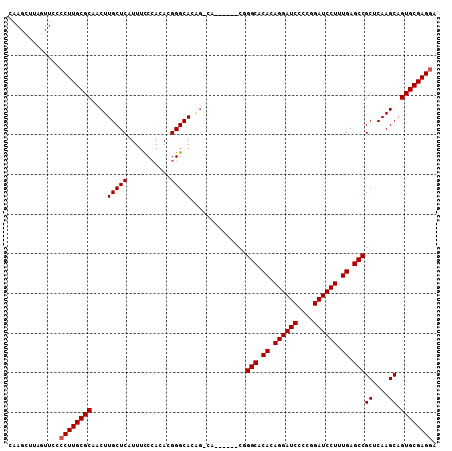
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:45:30 2006