| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,317,036 – 12,317,204 |
| Length | 168 |
| Max. P | 0.997195 |

| Location | 12,317,036 – 12,317,131 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 84.67 |
| Mean single sequence MFE | -26.58 |
| Consensus MFE | -21.20 |
| Energy contribution | -21.26 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.80 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.532759 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12317036 95 - 22224390 UUU------U----------AAUUUUGGCAAUAGCUGGUUGUCAUUAGUGGAGCAUGUCCAGGGCGUGAUUGAAUCUGAAUGGUAUAUUA------AGAAUCAGUUAUGGCUGCACG ...------.----------.....(((((((.....)))))))...(((.(((..((.....))((((((((.(((.((((...)))).------))).)))))))).))).))). ( -24.50) >DroSim_CAF1 19666 100 - 1 UUU------UUUU-----UUUUUUUUGGCAAUAGCUGGUUGUCAUUAGUGGAGCAUGUCCAGGGCGUGAUUGAAUCUGAAUGGAAUAUUA------AGAAUCAGUUAUGGCUGCACG ...------....-----.......(((((((.....)))))))...(((.(((..((.....))((((((((.(((.((((...)))).------))).)))))))).))).))). ( -24.50) >DroEre_CAF1 19598 117 - 1 UUGGCCACUUUUUUGGCCACUUUUUUGGCAAUAGCUGGUUGUCAUUAGUGGAGCAUGUCCAGGGCGUGAUUGAAUCUGAAUGGAAUAUUAAUAUUAGGAAUCAGUUAUGGCUGCACG .((((((......))))))......(((((((.....)))))))...(((.(((..((.....))((((((((.(((.((((.........)))).))).)))))))).))).))). ( -34.40) >DroYak_CAF1 18731 93 - 1 UU------------------UUAUUUGGCAAUAGCUGGUUGUCAUUAGUGGAGCAUGUCCAGGGCGUGAUUGAAUCUGAAUGGAAUAUUA------AGAAUCAGUUAUGGCUGCAGG ..------------------.....(((((((.....)))))))....((((.....))))((.(((((((((.(((.((((...)))).------))).))))))))).))..... ( -22.90) >consensus UUU______U__________UUUUUUGGCAAUAGCUGGUUGUCAUUAGUGGAGCAUGUCCAGGGCGUGAUUGAAUCUGAAUGGAAUAUUA______AGAAUCAGUUAUGGCUGCACG .........................(((((((.....)))))))...(((.(((..((.....))((((((((.(((...................))).)))))))).))).))). (-21.20 = -21.26 + 0.06)

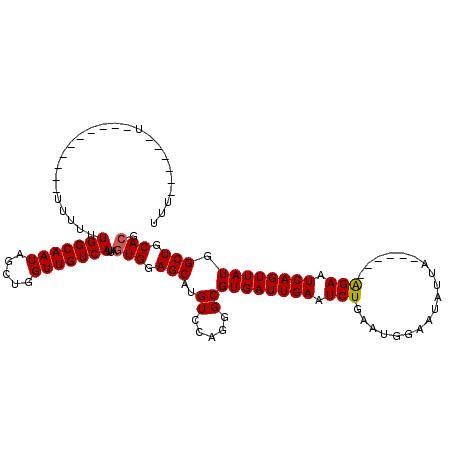

| Location | 12,317,107 – 12,317,204 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.76 |
| Mean single sequence MFE | -26.91 |
| Consensus MFE | -16.91 |
| Energy contribution | -17.13 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.63 |
| SVM decision value | 2.81 |
| SVM RNA-class probability | 0.997195 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12317107 97 + 22224390 AACCAGCUAUUGCCAAAAUU----------A------AAAAUAAGGGGCGUUGUUCGGCCACCUACAAGAAUAAUUAUAAUAAUAUUCCAGGAACGGGAAGGGUGGAGGGG--UA----- .(((..((.(..((......----------.------......((((((........))).))).....................((((......))))..))..))).))--).----- ( -20.00) >DroSim_CAF1 19737 92 + 1 AACCAGCUAUUGCCAAAAAAAA-----AAAA------AAAAUUAGGGGCGUUGUUCGGCCACCUACAAGAAUAAUUAUAAUAUUAUUCCAGGAGCGGGGGG------------UA----- .(((.(((.(((..........-----....------.....(((((((........))).))))...(((((((......)))))))))).)))....))------------).----- ( -20.70) >DroEre_CAF1 19675 120 + 1 AACCAGCUAUUGCCAAAAAAGUGGCCAAAAAAGUGGCCAAAUGAGGGGCGUUGUUCGGCCACCUACAAGCAUAAUUAUAAUAUUAUGCCAGGGCUGGGAGGGGCGGACGAUUCGAUAUGA ...........(((.......((((((......)))))).......)))((((((((.((.(((((..(((((((......)))))))....).))))..)).))))))))......... ( -40.04) >consensus AACCAGCUAUUGCCAAAAAA_______AAAA______AAAAUAAGGGGCGUUGUUCGGCCACCUACAAGAAUAAUUAUAAUAUUAUUCCAGGAACGGGAGGGG_GGA_G____UA_____ ..((..((....((.............................((((((........))).)))....(((((((......)))))))..))....))..)).................. (-16.91 = -17.13 + 0.22)


| Location | 12,317,107 – 12,317,204 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.76 |
| Mean single sequence MFE | -22.08 |
| Consensus MFE | -14.88 |
| Energy contribution | -14.77 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893704 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12317107 97 - 22224390 -----UA--CCCCUCCACCCUUCCCGUUCCUGGAAUAUUAUUAUAAUUAUUCUUGUAGGUGGCCGAACAACGCCCCUUAUUUU------U----------AAUUUUGGCAAUAGCUGGUU -----.(--((....((((.((((.......))))......(((((......)))))))))((((((................------.----------...)))))).......))). ( -13.75) >DroSim_CAF1 19737 92 - 1 -----UA------------CCCCCCGCUCCUGGAAUAAUAUUAUAAUUAUUCUUGUAGGUGGCCGAACAACGCCCCUAAUUUU------UUUU-----UUUUUUUUGGCAAUAGCUGGUU -----..------------...((.(((..(((((((((......)))))))...((((.(((........))))))).....------....-----..........))..))).)).. ( -18.70) >DroEre_CAF1 19675 120 - 1 UCAUAUCGAAUCGUCCGCCCCUCCCAGCCCUGGCAUAAUAUUAUAAUUAUGCUUGUAGGUGGCCGAACAACGCCCCUCAUUUGGCCACUUUUUUGGCCACUUUUUUGGCAAUAGCUGGUU .......................(((((...((((((((......))))))))...(((((((((((............))))))))))).....((((......))))....))))).. ( -33.80) >consensus _____UA____C_UCC_CCCCUCCCGCUCCUGGAAUAAUAUUAUAAUUAUUCUUGUAGGUGGCCGAACAACGCCCCUAAUUUU______UUUU_______UUUUUUGGCAAUAGCUGGUU ...............................((((((((......))))))))...(((.(((........))))))...........................(..((....))..).. (-14.88 = -14.77 + -0.11)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:32:15 2006