
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,303,942 – 12,304,057 |
| Length | 115 |
| Max. P | 0.999560 |

| Location | 12,303,942 – 12,304,057 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.50 |
| Mean single sequence MFE | -30.42 |
| Consensus MFE | -29.12 |
| Energy contribution | -29.32 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.58 |
| SVM RNA-class probability | 0.995494 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
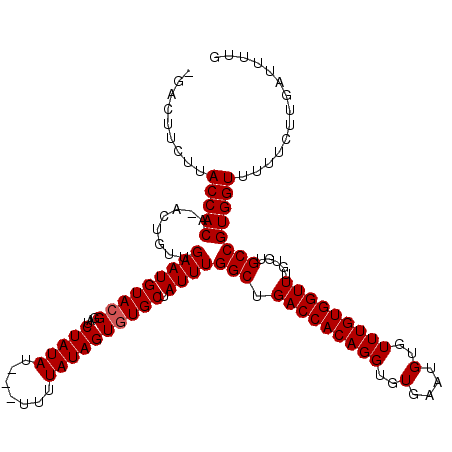
>X_DroMel_CAF1 12303942 115 + 22224390 -GACAUCUUACCACA-ACUGUUGAAUGUACGGAUCUAUAU---GUUUAUAGUGUGCUAUUUGGCUGACCACAGGUGUGAAUGUGUUUGUGGUUUGUGUGCCGUGGUUUUUCUUGAUUUUG -..((....(((((.-......(((((((((...(((((.---...)))))))))).))))(((.(((((((((..(....)..))))))))).....))))))))......))...... ( -30.70) >DroSec_CAF1 6200 117 + 1 GGACUUCUUACCACAAACUGUUGAAUGUACGGAUCUAUAU---UUUUAUAGUGUGCUAUUUGGCUGACCACAGGUGUGAAUGUGUUUGUGGUUUGUGUGCCGUGGUUUUUCUUGAUUUUG ((((..(..(((((((((.((..((((((((...(((((.---...)))))))))).)))..))....(((....))).....)))))))))..).)).))................... ( -30.30) >DroSim_CAF1 6725 116 + 1 -GACUGCUUACCACAAACUGUUGAAUGUACGGAUCUAUAU---UUUUAUAGUGUGCUAUUUGGCUGACCACAGGUGUGAAUGUGUUUGUGGUUUGUGUGCCGUGGUUUUUCUUGAUUUUG -(((..(....((((....((..((((((((...(((((.---...)))))))))).)))..)).(((((((((..(....)..)))))))))))))....)..)))............. ( -30.40) >DroEre_CAF1 6113 118 + 1 -GACUUCUCACCACA-ACUGUUGAAUGUAGGAAUCUAUAUUAUUUUUAUAGUGUGCUAUUUGGCUGACCACAGGUGUGAAUGUGUUUGUGGUUUGUGUGCCGUGGUUUUUCUUGAUUUUG -........(((((.-(((((..(((((((....)))))))......))))).........(((.(((((((((..(....)..))))))))).....)))))))).............. ( -30.40) >DroYak_CAF1 5582 118 + 1 -GACUUCUCACCACA-ACUGUUGAAUGUACGAGUCUAUAUUGUUUUUAUAGUGUGCUAUUUGGCUGACCACAGGUGUGAAUGUGUUUGUGGUUUGAGUGCCGUGGUUUUUCUUGAUUUUG -........(((((.-(((((..((((((((...(((((.......)))))))))).)))..)).(((((((((..(....)..)))))))))..)))...))))).............. ( -30.30) >consensus _GACUUCUUACCACA_ACUGUUGAAUGUACGGAUCUAUAU___UUUUAUAGUGUGCUAUUUGGCUGACCACAGGUGUGAAUGUGUUUGUGGUUUGUGUGCCGUGGUUUUUCUUGAUUUUG .........(((((........(((((((((...(((((.......)))))))))).))))(((.(((((((((..(....)..))))))))).....)))))))).............. (-29.12 = -29.32 + 0.20)

| Location | 12,303,942 – 12,304,057 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.50 |
| Mean single sequence MFE | -19.53 |
| Consensus MFE | -18.44 |
| Energy contribution | -18.44 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.72 |
| SVM RNA-class probability | 0.999560 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12303942 115 - 22224390 CAAAAUCAAGAAAAACCACGGCACACAAACCACAAACACAUUCACACCUGUGGUCAGCCAAAUAGCACACUAUAAAC---AUAUAGAUCCGUACAUUCAACAGU-UGUGGUAAGAUGUC- ..............((((((((......((((((..............))))))..((......))...(((((...---.)))))................))-))))))........- ( -20.04) >DroSec_CAF1 6200 117 - 1 CAAAAUCAAGAAAAACCACGGCACACAAACCACAAACACAUUCACACCUGUGGUCAGCCAAAUAGCACACUAUAAAA---AUAUAGAUCCGUACAUUCAACAGUUUGUGGUAAGAAGUCC ...................(((......(((((((((....((((....))))...((......))...(((((...---.)))))................))))))))).....))). ( -19.40) >DroSim_CAF1 6725 116 - 1 CAAAAUCAAGAAAAACCACGGCACACAAACCACAAACACAUUCACACCUGUGGUCAGCCAAAUAGCACACUAUAAAA---AUAUAGAUCCGUACAUUCAACAGUUUGUGGUAAGCAGUC- ..............((((((((......((((((..............))))))..)))..........(((((...---.)))))....................)))))........- ( -18.74) >DroEre_CAF1 6113 118 - 1 CAAAAUCAAGAAAAACCACGGCACACAAACCACAAACACAUUCACACCUGUGGUCAGCCAAAUAGCACACUAUAAAAAUAAUAUAGAUUCCUACAUUCAACAGU-UGUGGUGAGAAGUC- ..............((((((((......((((((..............))))))..((......))...(((((.......)))))................))-))))))........- ( -19.74) >DroYak_CAF1 5582 118 - 1 CAAAAUCAAGAAAAACCACGGCACUCAAACCACAAACACAUUCACACCUGUGGUCAGCCAAAUAGCACACUAUAAAAACAAUAUAGACUCGUACAUUCAACAGU-UGUGGUGAGAAGUC- ..............((((((((......((((((..............))))))..((......))...(((((.......)))))................))-))))))........- ( -19.74) >consensus CAAAAUCAAGAAAAACCACGGCACACAAACCACAAACACAUUCACACCUGUGGUCAGCCAAAUAGCACACUAUAAAA___AUAUAGAUCCGUACAUUCAACAGU_UGUGGUAAGAAGUC_ ..............((((((((......((((((..............))))))..)))..........(((((.......)))))....................)))))......... (-18.44 = -18.44 + -0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:32:04 2006