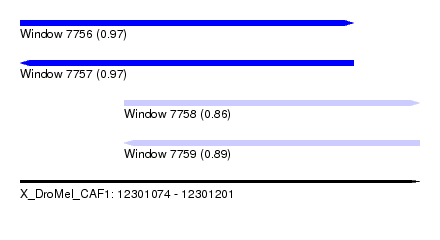
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,301,074 – 12,301,201 |
| Length | 127 |
| Max. P | 0.968218 |

| Location | 12,301,074 – 12,301,180 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 79.87 |
| Mean single sequence MFE | -29.15 |
| Consensus MFE | -18.24 |
| Energy contribution | -18.77 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965642 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12301074 106 + 22224390 AUGUACG----UGUGUCUGUGUGUGUGUGCAUUUUAAAGCAAUAUAUAUAUGUAUAUAAGUGGGUCAACUUCAUUCGGUGGUUGUUGACCCACAUCUUUUUCGGC-UGCAU (((((((----((((..(((((((((.(((........))))))))))))..)))))..((((((((((.((((...))))..))))))))))..........).-))))) ( -33.50) >DroVir_CAF1 1955 89 + 1 --GUG------UAUGUGUGUGUGUGUGUGCAUUUUAAGGCAUGAU--------------GUAGGUCAACUUCAUUUGGUGGUUGUUGACCCACAUCUUUUUGGGCCUGCUU --(((------((((..(....)..)))))))....((((..(((--------------((.(((((((.((((...))))..))))))).))))).......)))).... ( -27.70) >DroEre_CAF1 3229 102 + 1 ACGUAAG----UGUGUUUGUG--UGUGUGCAUUUUAAAGCAAUAU--AUAUAUAUAUAAGUGGGUCAACUUCAUUCGGUGGUUGUUGACCCACAUCUUUUUCGGC-UGCAU .((.(((----(((((.((((--(((((((........)).))))--))))).))))).((((((((((.((((...))))..))))))))))..)))...))..-..... ( -31.70) >DroWil_CAF1 6626 100 + 1 AUCUCCCUCUCUCUUUGUGUGUGUGUGUGCAUUUUAAACCAUAAU--GUUU--------GUAGGUCAACUUCAUUUGGUGGUUGUUGACCCACAUCUUUUUCGGU-UGCAU ................((((((((....))))....((((..((.--...(--------((.(((((((.((((...))))..))))))).)))....))..)))-))))) ( -19.50) >DroYak_CAF1 2747 100 + 1 AUGUAAG----UGUGUUUGUA--UGUGUGCAUUUUAAAGCAAUAU--AUA--UAUAUAAGUGGGUCAACUUCAUUCGGUGGUUGUUGACCCACAUCUUUUUCGGC-UGCAU (((((.(----(.(((.((((--(((((((........)).))))--)))--)).))).((((((((((.((((...))))..))))))))))..........))-))))) ( -31.00) >DroAna_CAF1 1549 102 + 1 AUGUACA----UAUGUAUGUGUAUGUGUGCAUUUUAAAGCAUUAU----GUGUAUAUAAGUGGGUCAACUUCAUUCGGUGGUUGAUGACCCACAUCUUUUUCGGG-UGCAC .(((((.----..((((((..((((.((((........)))))))----)..)))))).((((((((.(.((((...))))..).))))))))...........)-)))). ( -31.50) >consensus AUGUACG____UGUGUGUGUGUGUGUGUGCAUUUUAAAGCAAUAU__AUAU_UAUAUAAGUGGGUCAACUUCAUUCGGUGGUUGUUGACCCACAUCUUUUUCGGC_UGCAU ..........................(((((............................((((((((((.((((...))))..)))))))))).((......))..))))) (-18.24 = -18.77 + 0.53)

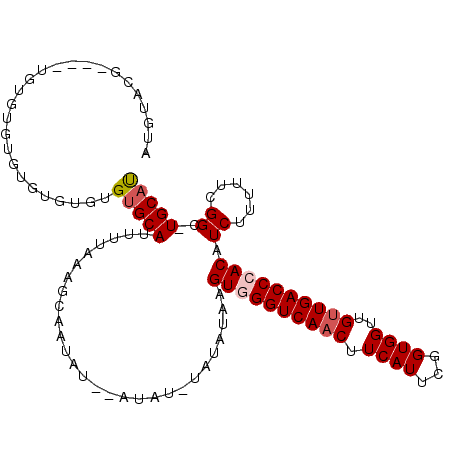
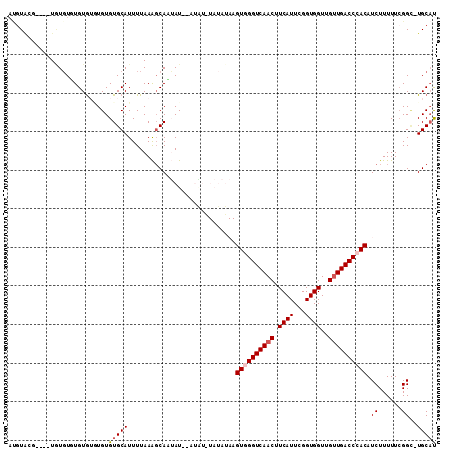
| Location | 12,301,074 – 12,301,180 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 79.87 |
| Mean single sequence MFE | -21.21 |
| Consensus MFE | -15.85 |
| Energy contribution | -15.97 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.09 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.968218 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12301074 106 - 22224390 AUGCA-GCCGAAAAAGAUGUGGGUCAACAACCACCGAAUGAAGUUGACCCACUUAUAUACAUAUAUAUAUUGCUUUAAAAUGCACACACACACAGACACA----CGUACAU .((((-......((((..((((((((((..(........)..)))))))))).((((((....))))))...))))....))))................----....... ( -22.20) >DroVir_CAF1 1955 89 - 1 AAGCAGGCCCAAAAAGAUGUGGGUCAACAACCACCAAAUGAAGUUGACCUAC--------------AUCAUGCCUUAAAAUGCACACACACACACACAUA------CAC-- ..((((((.......(((((((((((((..(........)..))))))))))--------------)))..)))......))).................------...-- ( -23.00) >DroEre_CAF1 3229 102 - 1 AUGCA-GCCGAAAAAGAUGUGGGUCAACAACCACCGAAUGAAGUUGACCCACUUAUAUAUAUAU--AUAUUGCUUUAAAAUGCACACA--CACAAACACA----CUUACGU .((((-......((((..((((((((((..(........)..)))))))))).(((((....))--)))...))))....))))....--..........----....... ( -21.30) >DroWil_CAF1 6626 100 - 1 AUGCA-ACCGAAAAAGAUGUGGGUCAACAACCACCAAAUGAAGUUGACCUAC--------AAAC--AUUAUGGUUUAAAAUGCACACACACACACAAAGAGAGAGGGAGAU .((((-((((.......(((((((((((..(........)..))))))))))--------)...--....))))......))))........................... ( -20.34) >DroYak_CAF1 2747 100 - 1 AUGCA-GCCGAAAAAGAUGUGGGUCAACAACCACCGAAUGAAGUUGACCCACUUAUAUA--UAU--AUAUUGCUUUAAAAUGCACACA--UACAAACACA----CUUACAU .((((-......((((..((((((((((..(........)..)))))))))).((((..--..)--)))...))))....))))....--..........----....... ( -20.00) >DroAna_CAF1 1549 102 - 1 GUGCA-CCCGAAAAAGAUGUGGGUCAUCAACCACCGAAUGAAGUUGACCCACUUAUAUACAC----AUAAUGCUUUAAAAUGCACACAUACACAUACAUA----UGUACAU (((((-............(((((((((((.........)))...))))))))..........----....(((........)))................----))))).. ( -20.40) >consensus AUGCA_GCCGAAAAAGAUGUGGGUCAACAACCACCGAAUGAAGUUGACCCACUUAUAUA_AUAU__AUAAUGCUUUAAAAUGCACACACACACAAACACA____CGUACAU ..................((((((((((..(........)..))))))))))..................(((........)))........................... (-15.85 = -15.97 + 0.11)



| Location | 12,301,107 – 12,301,201 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 80.56 |
| Mean single sequence MFE | -22.54 |
| Consensus MFE | -15.24 |
| Energy contribution | -14.85 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.862641 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
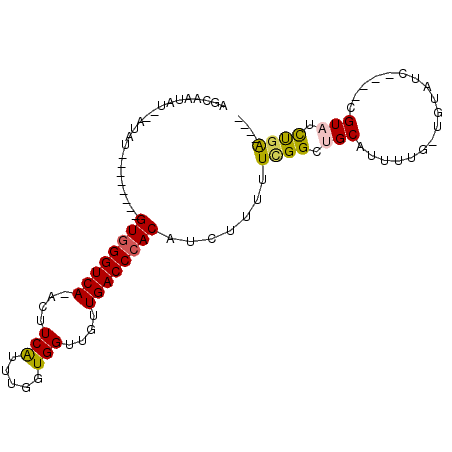
>X_DroMel_CAF1 12301107 94 + 22224390 AGCAAUAUAUAUAUGUAUAUAAGUGGGUCA-ACUUCAUUCGGUGGUUGUUGACCCACAUCUUUUUCGGCUGCAUUUUG-UGUAUCCC--CGUAUCUGA---- .....((((((....)))))).((((((((-((.((((...))))..)))))))))).((.....(((.(((((...)-))))...)--)).....))---- ( -25.30) >DroPse_CAF1 2502 82 + 1 AGCAAUAU--AAAU--------GUGGGUCAAACUUCGUUUGGUGGUUGUUGACCCACAUCUUUUUUGGCUGCAUU-UG-UGUAUC----UGUCUUCGG---- (((.....--..((--------(((((((((...(((.....)))...)))))))))))........))).....-..-......----.........---- ( -20.46) >DroSim_CAF1 3956 90 + 1 AGCAAUAU--AUAUGUAUAUAAGUGGGUCA-ACUUCAUUUGGUGGUUGUUGACCCACAUCUUUUUCGGCUGCAUUUUG-UGUAUC----CGUAUCUGA---- ....((((--(....)))))..((((((((-((.((((...))))..)))))))))).((.....(((.(((((...)-)))).)----)).....))---- ( -26.20) >DroWil_CAF1 6663 91 + 1 ACCAUAAU--GUUU--------GUAGGUCA-ACUUCAUUUGGUGGUUGUUGACCCACAUCUUUUUCGGUUGCAUUUCGUUGUAUCACUUCGUAUCUAAUAUA (((..((.--...(--------((.(((((-((.((((...))))..))))))).)))....))..)))((((......))))................... ( -16.90) >DroYak_CAF1 2778 88 + 1 AGCAAUAU--AUA--UAUAUAAGUGGGUCA-ACUUCAUUCGGUGGUUGUUGACCCACAUCUUUUUCGGCUGCAUUUUG-UGUAUC----CGUAUCUGA---- ........--...--.......((((((((-((.((((...))))..)))))))))).((.....(((.(((((...)-)))).)----)).....))---- ( -25.90) >DroPer_CAF1 2444 82 + 1 AGCAAUAU--AAAU--------GUGGGUCAAACUUCGUUUGGUGGUUGUUGACCCACAUCUUUUUUGGCUGCAUU-UG-UGUAUC----UGUCUUCGG---- (((.....--..((--------(((((((((...(((.....)))...)))))))))))........))).....-..-......----.........---- ( -20.46) >consensus AGCAAUAU__AUAU________GUGGGUCA_ACUUCAUUUGGUGGUUGUUGACCCACAUCUUUUUCGGCUGCAUUUUG_UGUAUC____CGUAUCUGA____ ......................((((((((....(((.....)))....)))))))).......((((.(((..................))).)))).... (-15.24 = -14.85 + -0.39)
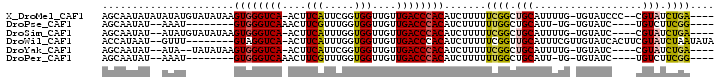

| Location | 12,301,107 – 12,301,201 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 80.56 |
| Mean single sequence MFE | -19.01 |
| Consensus MFE | -11.99 |
| Energy contribution | -11.85 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892352 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12301107 94 - 22224390 ----UCAGAUACG--GGGAUACA-CAAAAUGCAGCCGAAAAAGAUGUGGGUCAACAACCACCGAAUGAAGU-UGACCCACUUAUAUACAUAUAUAUAUUGCU ----.........--........-......((((...........((((((((((..(........)..))-)))))))).((((((....)))))))))). ( -20.50) >DroPse_CAF1 2502 82 - 1 ----CCGAAGACA----GAUACA-CA-AAUGCAGCCAAAAAAGAUGUGGGUCAACAACCACCAAACGAAGUUUGACCCAC--------AUUU--AUAUUGCU ----.........----......-..-.....(((......((((((((((((((..(........)..).)))))))))--------))))--.....))) ( -18.30) >DroSim_CAF1 3956 90 - 1 ----UCAGAUACG----GAUACA-CAAAAUGCAGCCGAAAAAGAUGUGGGUCAACAACCACCAAAUGAAGU-UGACCCACUUAUAUACAUAU--AUAUUGCU ----.......((----(...((-.....))...)))........((((((((((..(........)..))-))))))))............--........ ( -19.60) >DroWil_CAF1 6663 91 - 1 UAUAUUAGAUACGAAGUGAUACAACGAAAUGCAACCGAAAAAGAUGUGGGUCAACAACCACCAAAUGAAGU-UGACCUAC--------AAAC--AUUAUGGU ...............((......))........((((.......(((((((((((..(........)..))-))))))))--------)...--....)))) ( -17.74) >DroYak_CAF1 2778 88 - 1 ----UCAGAUACG----GAUACA-CAAAAUGCAGCCGAAAAAGAUGUGGGUCAACAACCACCGAAUGAAGU-UGACCCACUUAUAUA--UAU--AUAUUGCU ----.......((----(...((-.....))...)))........((((((((((..(........)..))-)))))))).......--...--........ ( -19.60) >DroPer_CAF1 2444 82 - 1 ----CCGAAGACA----GAUACA-CA-AAUGCAGCCAAAAAAGAUGUGGGUCAACAACCACCAAACGAAGUUUGACCCAC--------AUUU--AUAUUGCU ----.........----......-..-.....(((......((((((((((((((..(........)..).)))))))))--------))))--.....))) ( -18.30) >consensus ____UCAGAUACG____GAUACA_CAAAAUGCAGCCGAAAAAGAUGUGGGUCAACAACCACCAAAUGAAGU_UGACCCAC________AUAU__AUAUUGCU .............................................((((((((((..(........)..)).))))))))...................... (-11.99 = -11.85 + -0.14)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:32:00 2006