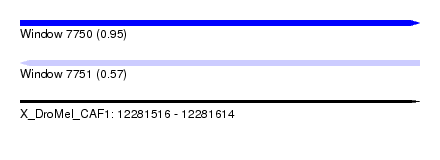
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,281,516 – 12,281,614 |
| Length | 98 |
| Max. P | 0.953048 |

| Location | 12,281,516 – 12,281,614 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 65.10 |
| Mean single sequence MFE | -39.67 |
| Consensus MFE | -17.98 |
| Energy contribution | -22.23 |
| Covariance contribution | 4.25 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.45 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953048 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
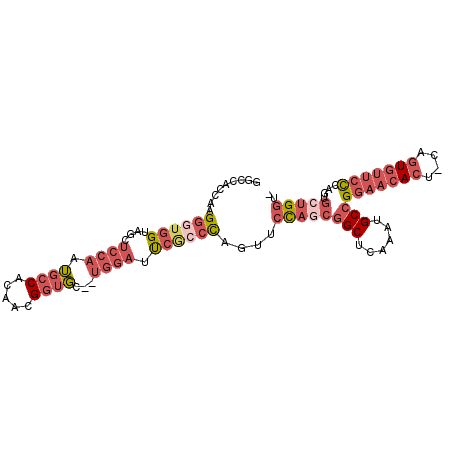
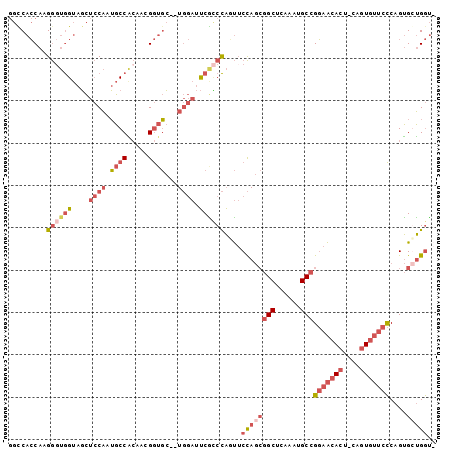
>X_DroMel_CAF1 12281516 98 + 22224390 GGUCACCAAGGCUGGUAGCUCCAAUGCCACAACGGUGC--UGGAUUCAUCCAAUUCCAGCGGCUCAAAUGCCGGAACACU-GAGUGUUCCCAGUGCUGGUA .(..((((....))))..)((((..(((.....)))..--))))...........((((((((......)))(((((((.-..)))))))....))))).. ( -35.80) >DroPse_CAF1 43227 88 + 1 --CCAGCAAGAGAAGAACUGACGAGGCCG-AUGGAACGAC----ACCGGCUGCCGCU----GCU-GGGGGCAGCAGCAGUACAGUCACCUCAGCGGCGAC- --...((..(((..((.((((((.(((((-.((......)----).)))))..)(((----(((-(....))))))).)).)))))..)))....))...- ( -32.70) >DroSec_CAF1 26790 98 + 1 GGCCACCAAGGGUGGUAGCUCCAAUGCCACAACGGUGC--UGGAUUCGCCCAGUUCCAGCGGCUCAAAUGCCGGAACACU-GGGUGUUCCCAGUGCUGGUA .((((((...)))))).((......)).....(((..(--(((...((((((((.....((((......))))....)))-)))))...))))..)))... ( -44.70) >DroEre_CAF1 24500 97 + 1 GGUCACCAAGGGUGGUAGCUCCAGUGCCGCAACGGUGC--UGGAUUCGCCCAGUUCCAGCGGCUCGAAUGCCGGAACACU-CAGUGUUCCCAGUGGUGGU- ..((((((.((((((....(((((..(((...)))..)--)))).)))))).........(((......)))(((((((.-..)))))))...)))))).- ( -45.50) >consensus GGCCACCAAGGGUGGUAGCUCCAAUGCCACAACGGUGC__UGGAUUCGCCCAGUUCCAGCGGCUCAAAUGCCGGAACACU_CAGUGUUCCCAGUGCUGGU_ .........((((((....((((.((((.....))))...)))).))))))....((((((((......)))(((((((....)))))))....))))).. (-17.98 = -22.23 + 4.25)

| Location | 12,281,516 – 12,281,614 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 65.10 |
| Mean single sequence MFE | -39.80 |
| Consensus MFE | -14.76 |
| Energy contribution | -17.82 |
| Covariance contribution | 3.06 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.37 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.572261 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
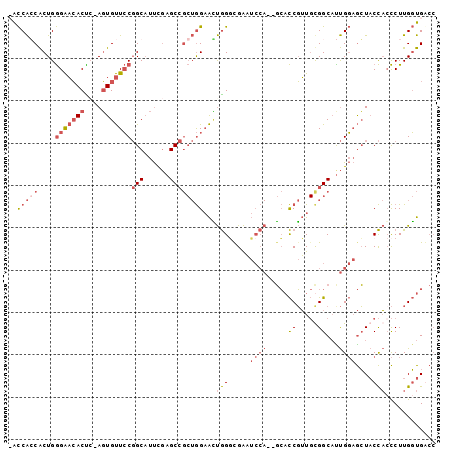
>X_DroMel_CAF1 12281516 98 - 22224390 UACCAGCACUGGGAACACUC-AGUGUUCCGGCAUUUGAGCCGCUGGAAUUGGAUGAAUCCA--GCACCGUUGUGGCAUUGGAGCUACCAGCCUUGGUGACC ..(((((....(((((((..-.)))))))(((......))))))))..(((((....))))--)(((((....(((..(((.....)))))).)))))... ( -39.20) >DroPse_CAF1 43227 88 - 1 -GUCGCCGCUGAGGUGACUGUACUGCUGCUGCCCCC-AGC----AGCGGCAGCCGGU----GUCGUUCCAU-CGGCCUCGUCAGUUCUUCUCUUGCUGG-- -....((((.((((.(((((.(((((((((((....-.))----)))))))((((((----(......)))-))))...)))))))..))))..)).))-- ( -39.40) >DroSec_CAF1 26790 98 - 1 UACCAGCACUGGGAACACCC-AGUGUUCCGGCAUUUGAGCCGCUGGAACUGGGCGAAUCCA--GCACCGUUGUGGCAUUGGAGCUACCACCCUUGGUGGCC .....((((((((....)))-)))))((((((......)))((((.(((.((((.......--)).))))).))))...)))(((((((....))))))). ( -42.60) >DroEre_CAF1 24500 97 - 1 -ACCACCACUGGGAACACUG-AGUGUUCCGGCAUUCGAGCCGCUGGAACUGGGCGAAUCCA--GCACCGUUGCGGCACUGGAGCUACCACCCUUGGUGACC -..(((((..(((.....((-.((((((((((......)))((((.(((.((((.......--)).))))).))))...))))).))))))).)))))... ( -38.00) >consensus _ACCACCACUGGGAACACUC_AGUGUUCCGGCAUUCGAGCCGCUGGAACUGGGCGAAUCCA__GCACCGUUGCGGCAUUGGAGCUACCACCCUUGGUGACC ..(((((....(((((((....)))))))(((......))))))))...(((.....((((.....((.....))...))))....)))............ (-14.76 = -17.82 + 3.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:31:53 2006