
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,259,022 – 12,259,131 |
| Length | 109 |
| Max. P | 0.942029 |

| Location | 12,259,022 – 12,259,131 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.88 |
| Mean single sequence MFE | -17.50 |
| Consensus MFE | -16.20 |
| Energy contribution | -16.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.942029 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
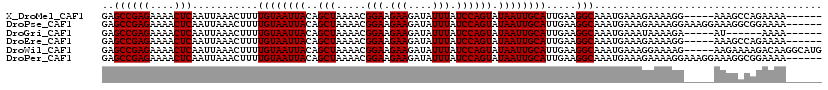
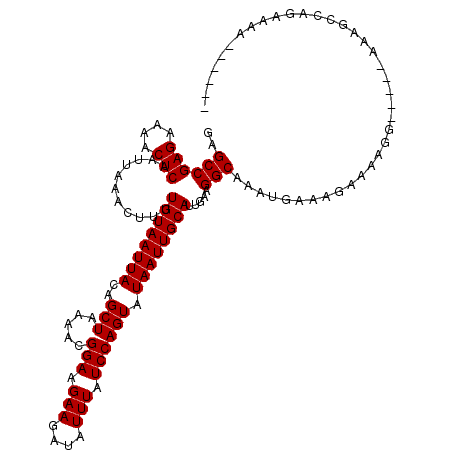
>X_DroMel_CAF1 12259022 109 + 22224390 GAGCCGAGAAAACUCAAUUAAACUUUUGUAAUUACAGCUAAAACGGAAGAAGAUAUUUAUCCAGUAUAAUUGCAUUGAAGGCAAAUGAAAGAAAAGG-----AAAGCCAGAAAA------ ..((((((....)))...........((((((((..(((.....(((.(((....))).)))))).)))))))).....))).............((-----....))......------ ( -17.10) >DroPse_CAF1 15606 114 + 1 GAGCCGAGAAAACUCAAUUAAACUUUUGUAAUUACAGCUAAAACGGAAGAAGAUAUUUAUCCAGUAUAAUUGCAUUGAAGGCAAAUGAAAGAAAAGGAAAGGAAAGGCGGAAAA------ ..((((((....))).......((((((((((((..(((.....(((.(((....))).)))))).))))))))..)))).........................)))......------ ( -18.20) >DroGri_CAF1 6404 103 + 1 GAGCCGAGAAAACUCAAUUAAACUUUUGUAAUUACAGCUAAAACGGAAGAAGAUAUUUAUCCAGUAUAAUUGCAUUGAAGGCAAAUGAAAUAAAAGA-----AU------AAAA------ ..((((((....)))...........((((((((..(((.....(((.(((....))).)))))).)))))))).....)))...............-----..------....------ ( -16.20) >DroEre_CAF1 3621 109 + 1 GAGCCGAGAAAACUCAAUUAAACUUUUGUAAUUACAGCUAAAACGGAAGAAGAUAUUUAUCCAGUAUAAUUGCAUUGAAGGCAAAUGAAAGAAAAGG-----AAAGCCAGAAAA------ ..((((((....)))...........((((((((..(((.....(((.(((....))).)))))).)))))))).....))).............((-----....))......------ ( -17.10) >DroWil_CAF1 4910 115 + 1 GAGCCGAGAAAACUCAAUUAAACUUUUGUAAUUACAGCUAAAACGGAAGAAGAUAUUUAUCCAGUAUAAUUGCAUUGAAGGCAAAUGAAAGGAAAAG-----AAGAAAAGACAAGGCAUG ..((((((....))).......((((((((((((..(((.....(((.(((....))).)))))).))))))))..)))).................-----............)))... ( -18.20) >DroPer_CAF1 14613 114 + 1 GAGCCGAGAAAACUCAAUUAAACUUUUGUAAUUACAGCUAAAACGGAAGAAGAUAUUUAUCCAGUAUAAUUGCAUUGAAGGCAAAUGAAAGAAAAGGAAAGGAAAGGCGGAAAA------ ..((((((....))).......((((((((((((..(((.....(((.(((....))).)))))).))))))))..)))).........................)))......------ ( -18.20) >consensus GAGCCGAGAAAACUCAAUUAAACUUUUGUAAUUACAGCUAAAACGGAAGAAGAUAUUUAUCCAGUAUAAUUGCAUUGAAGGCAAAUGAAAGAAAAGG_____AAAGCCAGAAAA______ ..((((((....)))...........((((((((..(((.....(((.(((....))).)))))).)))))))).....)))...................................... (-16.20 = -16.20 + -0.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:31:43 2006