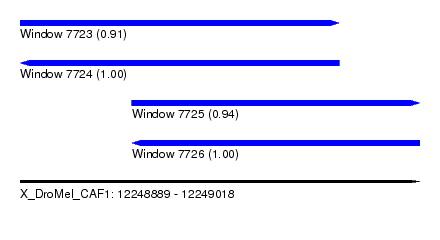
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,248,889 – 12,249,018 |
| Length | 129 |
| Max. P | 0.999167 |

| Location | 12,248,889 – 12,248,992 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.93 |
| Mean single sequence MFE | -31.86 |
| Consensus MFE | -26.74 |
| Energy contribution | -26.62 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.906000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12248889 103 + 22224390 ----CAGAGAGAGUAGUAGUUUGUCAAAAGUGCGACUACAAAAGCAC------UCGCUGCUUAUCGCACCAGACAGACUUUUCCGACGGUCGGAAAAGUCCAUGG-------GGGGAAAU ----......(((((((.(.....)...(((((..........))))------).))))))).((.(.(((....((((((((((.....))))))))))..)))-------.).))... ( -32.20) >DroSec_CAF1 16944 102 + 1 ----CAGAGAGAGUAGUAGUUUGUCAAAAGUGCGACUACAAAAGCAC------UCCCUGCUUAUCGCACCAGACAGACUUUUCCGACGGUCGGAAAAGUCCAUGG-------G-GGAAAU ----..((.(((.......))).))....((((..........))))------(((((((.....)))(((....((((((((((.....))))))))))..)))-------)-)))... ( -31.70) >DroSim_CAF1 6729 102 + 1 ----CAGAGAGAGUAGUAGUUUGUCAAAAGUGCGACUACAAAAGCAC------UCGCUGCUUAUCGCACCAGACAGACUUUUCCGACGGUCGGAAAAGUCCAUGG-------G-GGAAAU ----......(((((((.(.....)...(((((..........))))------).))))))).((.(.(((....((((((((((.....))))))))))..)))-------)-.))... ( -32.10) >DroEre_CAF1 12047 101 + 1 ----AAGAGAGAGUUGUAGUUUGUCAAAAGUGCGACUACAAAAGUGCGACUGCUCGCUGCUUAUCGCACCAGACAGACAUUUCCGACGGUCGGAAAAGUCGGCGG--------------- ----.......(((((((.(((....))).)))))))......((((((..((.....))...))))))......(((.((((((.....)))))).))).....--------------- ( -31.90) >DroYak_CAF1 5307 111 + 1 CACACAGAGAGAG---UAGUUUGUCAAAAGUGCGACUACAAAAGCAC-----UCCGCUGCUUAUCGCACCAGACAGACUUUUCCGACGGUCGGAAAAGUCCGCAGUCCAUAGG-GAAAAU ............(---(....((((....((((((.......(((..-----...))).....))))))..))))((((((((((.....)))))))))).))..(((....)-)).... ( -31.40) >consensus ____CAGAGAGAGUAGUAGUUUGUCAAAAGUGCGACUACAAAAGCAC______UCGCUGCUUAUCGCACCAGACAGACUUUUCCGACGGUCGGAAAAGUCCAUGG_______G_GGAAAU .....................((((....((((((......(((((...........))))).))))))..))))((((((((((.....)))))))))).................... (-26.74 = -26.62 + -0.12)

| Location | 12,248,889 – 12,248,992 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.93 |
| Mean single sequence MFE | -34.96 |
| Consensus MFE | -31.12 |
| Energy contribution | -31.72 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.89 |
| SVM decision value | 3.41 |
| SVM RNA-class probability | 0.999167 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12248889 103 - 22224390 AUUUCCCC-------CCAUGGACUUUUCCGACCGUCGGAAAAGUCUGUCUGGUGCGAUAAGCAGCGA------GUGCUUUUGUAGUCGCACUUUUGACAAACUACUACUCUCUCUG---- ........-------....(((((((((((.....)))))))))))(((.(((((((((((((....------.))))).....))))))))...)))..................---- ( -34.90) >DroSec_CAF1 16944 102 - 1 AUUUCC-C-------CCAUGGACUUUUCCGACCGUCGGAAAAGUCUGUCUGGUGCGAUAAGCAGGGA------GUGCUUUUGUAGUCGCACUUUUGACAAACUACUACUCUCUCUG---- ......-.-------....(((((((((((.....)))))))))))(((.(((((((((((((....------.))))).....))))))))...)))..................---- ( -34.90) >DroSim_CAF1 6729 102 - 1 AUUUCC-C-------CCAUGGACUUUUCCGACCGUCGGAAAAGUCUGUCUGGUGCGAUAAGCAGCGA------GUGCUUUUGUAGUCGCACUUUUGACAAACUACUACUCUCUCUG---- ......-.-------....(((((((((((.....)))))))))))(((.(((((((((((((....------.))))).....))))))))...)))..................---- ( -34.90) >DroEre_CAF1 12047 101 - 1 ---------------CCGCCGACUUUUCCGACCGUCGGAAAUGUCUGUCUGGUGCGAUAAGCAGCGAGCAGUCGCACUUUUGUAGUCGCACUUUUGACAAACUACAACUCUCUCUU---- ---------------.....(((.((((((.....)))))).)))((((.((((((((.....((((....))))((....)).))))))))...)))).................---- ( -32.20) >DroYak_CAF1 5307 111 - 1 AUUUUC-CCUAUGGACUGCGGACUUUUCCGACCGUCGGAAAAGUCUGUCUGGUGCGAUAAGCAGCGGA-----GUGCUUUUGUAGUCGCACUUUUGACAAACUA---CUCUCUCUGUGUG ......-......(((((((((((((((((.....))))))))))).((((.(((.....))).))))-----........))))))((((....((.......---.)).....)))). ( -37.90) >consensus AUUUCC_C_______CCAUGGACUUUUCCGACCGUCGGAAAAGUCUGUCUGGUGCGAUAAGCAGCGA______GUGCUUUUGUAGUCGCACUUUUGACAAACUACUACUCUCUCUG____ ....................((((((((((.....))))))))))((((.(((((((((((((...........))))).....))))))))...))))..................... (-31.12 = -31.72 + 0.60)


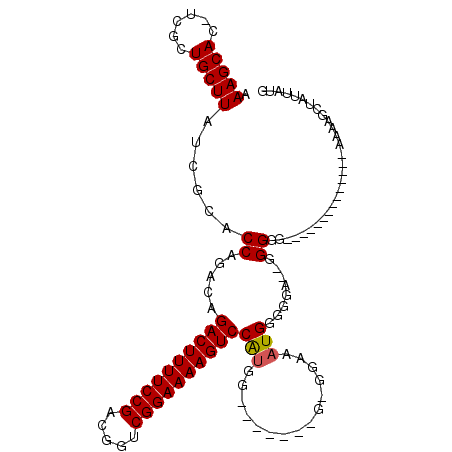
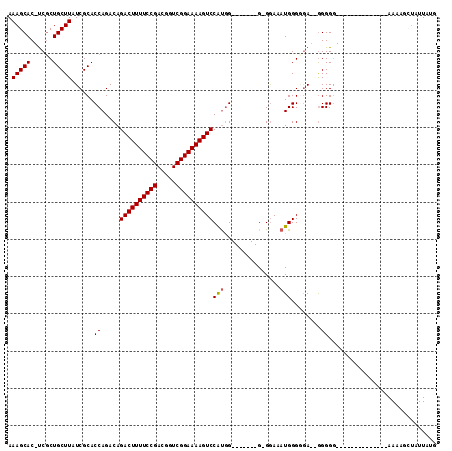
| Location | 12,248,925 – 12,249,018 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 79.48 |
| Mean single sequence MFE | -30.89 |
| Consensus MFE | -19.45 |
| Energy contribution | -19.51 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938817 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12248925 93 + 22224390 AAAGCAC-UCGCUGCUUAUCGCACCAGACAGACUUUUCCGACGGUCGGAAAAGUCCAUGG-------GGGGAAAUGGGUGAUGGGGGG--------------AAAAGCUAUUAUG ..(((..-...((.((((((((.(((..((((((((((((.....))))))))))..)).-------.......))))))))))).))--------------....)))...... ( -32.00) >DroSec_CAF1 16980 90 + 1 AAAGCAC-UCCCUGCUUAUCGCACCAGACAGACUUUUCCGACGGUCGGAAAAGUCCAUGG-------G-GGAAAUGGGGGA--AGGGG--------------AAAGGAUAUUAUG .......-(((((.(((((..(.(((....((((((((((.....))))))))))..)))-------.-.)..)))))...--.))))--------------)............ ( -29.40) >DroSim_CAF1 6765 90 + 1 AAAGCAC-UCGCUGCUUAUCGCACCAGACAGACUUUUCCGACGGUCGGAAAAGUCCAUGG-------G-GGAAAUGGGGGA--GGGGG--------------AAAAGCUAUUAUG ..(((.(-((.((.(((.((.(.(((....((((((((((.....))))))))))..)))-------)-.))....))).)--).)))--------------....)))...... ( -30.50) >DroYak_CAF1 5344 110 + 1 AAAGCACUCCGCUGCUUAUCGCACCAGACAGACUUUUCCGACGGUCGGAAAAGUCCGCAGUCCAUAGG-GAAAAUGGGAAAAAGGGGAAAAAGGGGGUAGGGAAA----AUUAUG .......(((.((((((......((.....((((((((((.....))))))))))(.((.(((....)-))...)).)......))........)))))))))..----...... ( -31.64) >consensus AAAGCAC_UCGCUGCUUAUCGCACCAGACAGACUUUUCCGACGGUCGGAAAAGUCCAUGG_______G_GGAAAUGGGGGA__GGGGG______________AAAAGCUAUUAUG .(((((......)))))......((.....((((((((((.....))))))))))(((...............)))........))............................. (-19.45 = -19.51 + 0.06)

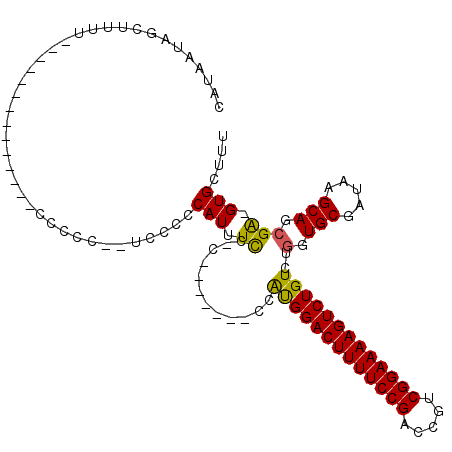
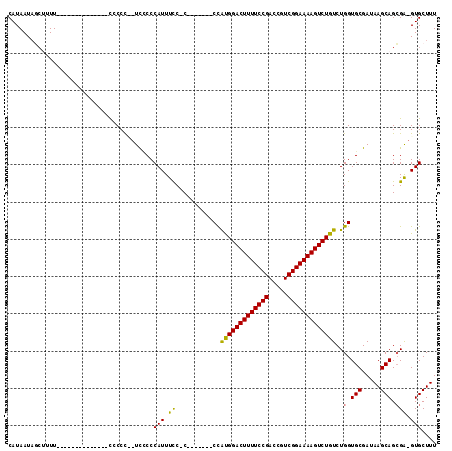
| Location | 12,248,925 – 12,249,018 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 79.48 |
| Mean single sequence MFE | -27.25 |
| Consensus MFE | -22.40 |
| Energy contribution | -21.90 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.79 |
| SVM RNA-class probability | 0.997071 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12248925 93 - 22224390 CAUAAUAGCUUUU--------------CCCCCCAUCACCCAUUUCCCC-------CCAUGGACUUUUCCGACCGUCGGAAAAGUCUGUCUGGUGCGAUAAGCAGCGA-GUGCUUU ......(((..((--------------(....................-------..(((((((((((((.....)))))))))))))..(.(((.....))).)))-).))).. ( -24.70) >DroSec_CAF1 16980 90 - 1 CAUAAUAUCCUUU--------------CCCCU--UCCCCCAUUUCC-C-------CCAUGGACUUUUCCGACCGUCGGAAAAGUCUGUCUGGUGCGAUAAGCAGGGA-GUGCUUU .............--------------.....--.....(((((((-(-------(.(((((((((((((.....)))))))))))))..))(((.....)))))))-))).... ( -28.30) >DroSim_CAF1 6765 90 - 1 CAUAAUAGCUUUU--------------CCCCC--UCCCCCAUUUCC-C-------CCAUGGACUUUUCCGACCGUCGGAAAAGUCUGUCUGGUGCGAUAAGCAGCGA-GUGCUUU ......(((..((--------------(....--............-.-------..(((((((((((((.....)))))))))))))..(.(((.....))).)))-).))).. ( -24.70) >DroYak_CAF1 5344 110 - 1 CAUAAU----UUUCCCUACCCCCUUUUUCCCCUUUUUCCCAUUUUC-CCUAUGGACUGCGGACUUUUCCGACCGUCGGAAAAGUCUGUCUGGUGCGAUAAGCAGCGGAGUGCUUU ......----............................((((....-...))))((((((((((((((((.....)))))))))))))(((.(((.....))).))))))..... ( -31.30) >consensus CAUAAUAGCUUUU______________CCCCC__UCCCCCAUUUCC_C_______CCAUGGACUUUUCCGACCGUCGGAAAAGUCUGUCUGGUGCGAUAAGCAGCGA_GUGCUUU .......................................(((.((............(((((((((((((.....)))))))))))))..(.(((.....))).))).))).... (-22.40 = -21.90 + -0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:31:31 2006