
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,243,999 – 12,244,121 |
| Length | 122 |
| Max. P | 0.994369 |

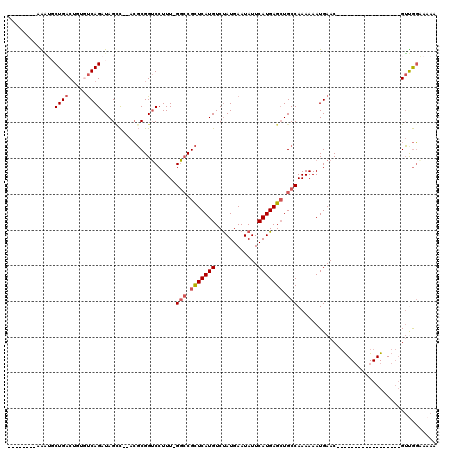
| Location | 12,243,999 – 12,244,092 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.89 |
| Mean single sequence MFE | -34.47 |
| Consensus MFE | -15.87 |
| Energy contribution | -16.40 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.46 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984468 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12243999 93 + 22224390 --------AAAUGCUGACUGUGUCAGAUAGGCCAACGCGGUCCUUU-GGCCGCUCAUGUCUAUGAAUAUUCAUGAGAUGCCAAAAAAUGAAC------------------GUUGGAAAAA --------.....(((((...))))).....((((((..((..(((-(((..((((((..((....))..))))))..))))))..))...)------------------)))))..... ( -27.40) >DroPse_CAF1 43338 109 + 1 --------CGGGUCUGACUCGCUCAGAUAGCC--ACGCGGUCCUUU-GGCCGCUCAUGUCUAUGAAUAUUCAUGAGCUGCCAAAAAAUGAACCCCGCCAGAGGCGUCGGCGUCGGUAAAA --------...((((((.....)))))).(((--(((((((.((((-(((.(((((((..((....))..))))))).))))))....).))).((((...))))...)))).))).... ( -39.60) >DroGri_CAF1 13076 115 + 1 GGCUGAGGGGCUGCUGACGUCGCCAGAUAGCU--AGCCGGGCCUUUAGUGCGUUCAUGUCCAUGAAUAUUCAUGAGCUGCCAAAAAAUGAACCGUGCAUAAGGCGAC---GGUACCAAAA (((((...(((.((....)).)))...)))))--.((((.(((((..(..((((((((..(((((....)))))..).........))))).))..)..)))))..)---)))....... ( -37.80) >DroEre_CAF1 8932 93 + 1 --------AAAUGCUGACUGUGUCAGAUAGUCAAACGCGGUCCUUU-GGCCGCUCAUGUCUAUGAAUAUUCAUGAGCUGCCAAAAAAUGAAC------------------GUUGGAAAAA --------.((((..((((((((..(.....)..)))))))).(((-(((.(((((((..((....))..))))))).)))))).......)------------------)))....... ( -29.40) >DroYak_CAF1 1301 93 + 1 --------AAAUGCUGACUGUGUCAGAUAGUCCAACGCGGUCCUUU-GGCCGCUCAUGUCUAUGAAUAUUCAUGAGCUGCCAAAAAAUGAAC------------------GUUGGAAAAA --------.....(((((...)))))....(((((((..((..(((-(((.(((((((..((....))..))))))).))))))..))...)------------------)))))).... ( -33.00) >DroPer_CAF1 40478 109 + 1 --------CAGGUCUGACUCGCUCAGAUAGCC--ACGCGGUCCUUU-GGCCGCUCAUGUCUAUGAAUAUUCAUGAGCUGCCAAAAAAUGAACCCCGCCAGAGGCGUCGGCGUCGGUAAAA --------...((((((.....)))))).(((--(((((((.((((-(((.(((((((..((....))..))))))).))))))....).))).((((...))))...)))).))).... ( -39.60) >consensus ________AAAUGCUGACUGUGUCAGAUAGCC__ACGCGGUCCUUU_GGCCGCUCAUGUCUAUGAAUAUUCAUGAGCUGCCAAAAAAUGAAC__________________GUUGGAAAAA .............((((.....)))).....................(((.(((((((............))))))).)))....................................... (-15.87 = -16.40 + 0.53)


| Location | 12,243,999 – 12,244,092 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.89 |
| Mean single sequence MFE | -34.65 |
| Consensus MFE | -16.54 |
| Energy contribution | -17.10 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.48 |
| SVM decision value | 2.47 |
| SVM RNA-class probability | 0.994369 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
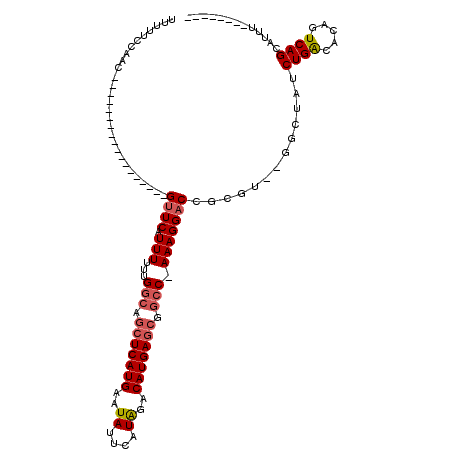
>X_DroMel_CAF1 12243999 93 - 22224390 UUUUUCCAAC------------------GUUCAUUUUUUGGCAUCUCAUGAAUAUUCAUAGACAUGAGCGGCC-AAAGGACCGCGUUGGCCUAUCUGACACAGUCAGCAUUU-------- .....(((((------------------((...(((((((((..((((((..((....))..))))))..)))-))))))..))))))).....(((((...))))).....-------- ( -30.50) >DroPse_CAF1 43338 109 - 1 UUUUACCGACGCCGACGCCUCUGGCGGGGUUCAUUUUUUGGCAGCUCAUGAAUAUUCAUAGACAUGAGCGGCC-AAAGGACCGCGU--GGCUAUCUGAGCGAGUCAGACCCG-------- .........(((((.......)))))(((((....(((((((.(((((((..((....))..))))))).)))-))))((((((..--((....))..))).))).))))).-------- ( -40.90) >DroGri_CAF1 13076 115 - 1 UUUUGGUACC---GUCGCCUUAUGCACGGUUCAUUUUUUGGCAGCUCAUGAAUAUUCAUGGACAUGAACGCACUAAAGGCCCGGCU--AGCUAUCUGGCGACGUCAGCAGCCCCUCAGCC ..(((.(..(---((((((((.(((...((((((.........(..(((((....)))))..))))))))))...)))))...(((--((....)))))))))..).))).......... ( -28.90) >DroEre_CAF1 8932 93 - 1 UUUUUCCAAC------------------GUUCAUUUUUUGGCAGCUCAUGAAUAUUCAUAGACAUGAGCGGCC-AAAGGACCGCGUUUGACUAUCUGACACAGUCAGCAUUU-------- ....((.(((------------------((...(((((((((.(((((((..((....))..))))))).)))-))))))..))))).))....(((((...))))).....-------- ( -30.30) >DroYak_CAF1 1301 93 - 1 UUUUUCCAAC------------------GUUCAUUUUUUGGCAGCUCAUGAAUAUUCAUAGACAUGAGCGGCC-AAAGGACCGCGUUGGACUAUCUGACACAGUCAGCAUUU-------- ....((((((------------------((...(((((((((.(((((((..((....))..))))))).)))-))))))..))))))))....(((((...))))).....-------- ( -36.80) >DroPer_CAF1 40478 109 - 1 UUUUACCGACGCCGACGCCUCUGGCGGGGUUCAUUUUUUGGCAGCUCAUGAAUAUUCAUAGACAUGAGCGGCC-AAAGGACCGCGU--GGCUAUCUGAGCGAGUCAGACCUG-------- .....((.((((...((((...)))).(((((....((((((.(((((((..((....))..))))))).)))-))))))))))))--))...(((((.....)))))....-------- ( -40.50) >consensus UUUUUCCAAC__________________GUUCAUUUUUUGGCAGCUCAUGAAUAUUCAUAGACAUGAGCGGCC_AAAGGACCGCGU__GGCUAUCUGACACAGUCAGCAUUU________ ............................((((.(((...(((.(((((((..((....))..))))))).))).))))))).............((((.....))))............. (-16.54 = -17.10 + 0.56)
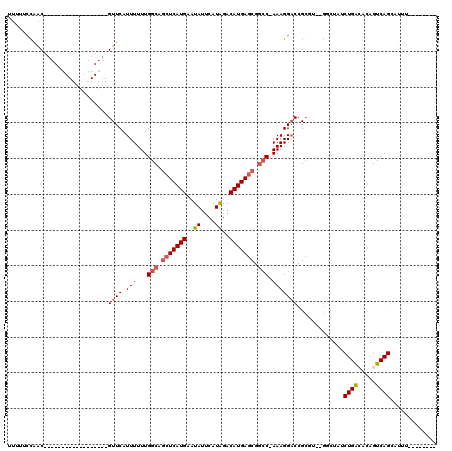
| Location | 12,244,031 – 12,244,121 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 79.51 |
| Mean single sequence MFE | -33.95 |
| Consensus MFE | -27.00 |
| Energy contribution | -26.64 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968584 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12244031 90 + 22224390 UCCUUUGGCCGCUCAUGUCUAUGAAUAUUCAUGAGAUGCCAAAAAAUGAAC------------------GUUGGAAAAACAUCCACGGCCCAAUGGA-------AACGUGGGUGG ...((((((..((((((..((....))..))))))..)))))).......(------------------((.(((......))))))(((((.((..-------..))))))).. ( -30.90) >DroPse_CAF1 43368 108 + 1 UCCUUUGGCCGCUCAUGUCUAUGAAUAUUCAUGAGCUGCCAAAAAAUGAACCCCGCCAGAGGCGUCGGCGUCGGUAAAACAUCAGAGAAGGAGAGAA-------GACUCGGGUGG ...((((((.(((((((..((....))..))))))).))))))......((((.(((.((.((....)).))))).................(((..-------..))))))).. ( -34.40) >DroSim_CAF1 1190 90 + 1 UCCUUUGGCCGCUCAUGUCUAUGAAUAUUCAUGAGAUGCCAAAAAAUGAAC------------------GUUGGAAAAACAUCCACGGCCCAAUGGA-------AGCGUGGGUGG (((((((((..((((((..((....))..))))))..)))))).(((....------------------))))))....((((((((.((....)).-------..)))))))). ( -29.70) >DroEre_CAF1 8964 90 + 1 UCCUUUGGCCGCUCAUGUCUAUGAAUAUUCAUGAGCUGCCAAAAAAUGAAC------------------GUUGGAAAAACAUCCACGGCCCAAUGGA-------AACGUGGGUGG ...((((((.(((((((..((....))..))))))).)))))).......(------------------((.(((......))))))(((((.((..-------..))))))).. ( -34.90) >DroYak_CAF1 1333 97 + 1 UCCUUUGGCCGCUCAUGUCUAUGAAUAUUCAUGAGCUGCCAAAAAAUGAAC------------------GUUGGAAAAACAUCCACGGCCCAAUGGACGUGGGGAACGUGGGUGG (((((((((.(((((((..((....))..))))))).)))))).(((....------------------))))))....((((((((.((((.......))))...)))))))). ( -38.30) >DroPer_CAF1 40508 108 + 1 UCCUUUGGCCGCUCAUGUCUAUGAAUAUUCAUGAGCUGCCAAAAAAUGAACCCCGCCAGAGGCGUCGGCGUCGGUAAAACGUCAGAGAAGGAGAGAA-------GACUCGGGUGG ...((((((.(((((((..((....))..))))))).))))))......((((((((...))))..(((((.......))))).........(((..-------..))))))).. ( -35.50) >consensus UCCUUUGGCCGCUCAUGUCUAUGAAUAUUCAUGAGCUGCCAAAAAAUGAAC__________________GUUGGAAAAACAUCCACGGCCCAAUGGA_______AACGUGGGUGG ...((((((.(((((((..((....))..))))))).))))))....................................((((((((...................)))))))). (-27.00 = -26.64 + -0.36)


| Location | 12,244,031 – 12,244,121 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 79.51 |
| Mean single sequence MFE | -31.30 |
| Consensus MFE | -23.05 |
| Energy contribution | -22.83 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963291 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12244031 90 - 22224390 CCACCCACGUU-------UCCAUUGGGCCGUGGAUGUUUUUCCAAC------------------GUUCAUUUUUUGGCAUCUCAUGAAUAUUCAUAGACAUGAGCGGCCAAAGGA ...((((....-------.....))))(((((((((((.....)))------------------))))))..((((((..((((((..((....))..))))))..)))))))). ( -26.70) >DroPse_CAF1 43368 108 - 1 CCACCCGAGUC-------UUCUCUCCUUCUCUGAUGUUUUACCGACGCCGACGCCUCUGGCGGGGUUCAUUUUUUGGCAGCUCAUGAAUAUUCAUAGACAUGAGCGGCCAAAGGA (((((((((..-------..)))......................(((((.......)))))))))......((((((.(((((((..((....))..))))))).)))))))). ( -35.30) >DroSim_CAF1 1190 90 - 1 CCACCCACGCU-------UCCAUUGGGCCGUGGAUGUUUUUCCAAC------------------GUUCAUUUUUUGGCAUCUCAUGAAUAUUCAUAGACAUGAGCGGCCAAAGGA ...((((....-------.....))))(((((((((((.....)))------------------))))))..((((((..((((((..((....))..))))))..)))))))). ( -26.70) >DroEre_CAF1 8964 90 - 1 CCACCCACGUU-------UCCAUUGGGCCGUGGAUGUUUUUCCAAC------------------GUUCAUUUUUUGGCAGCUCAUGAAUAUUCAUAGACAUGAGCGGCCAAAGGA ...((((....-------.....))))(((((((((((.....)))------------------))))))..((((((.(((((((..((....))..))))))).)))))))). ( -30.90) >DroYak_CAF1 1333 97 - 1 CCACCCACGUUCCCCACGUCCAUUGGGCCGUGGAUGUUUUUCCAAC------------------GUUCAUUUUUUGGCAGCUCAUGAAUAUUCAUAGACAUGAGCGGCCAAAGGA .((.(((((...((((.......)))).))))).))..........------------------.....(((((((((.(((((((..((....))..))))))).))))))))) ( -32.90) >DroPer_CAF1 40508 108 - 1 CCACCCGAGUC-------UUCUCUCCUUCUCUGACGUUUUACCGACGCCGACGCCUCUGGCGGGGUUCAUUUUUUGGCAGCUCAUGAAUAUUCAUAGACAUGAGCGGCCAAAGGA (((((((((..-------..)))......................(((((.......)))))))))......((((((.(((((((..((....))..))))))).)))))))). ( -35.30) >consensus CCACCCACGUU_______UCCAUUGGGCCGUGGAUGUUUUUCCAAC__________________GUUCAUUUUUUGGCAGCUCAUGAAUAUUCAUAGACAUGAGCGGCCAAAGGA ((..(((((...................))))).......................................((((((.(((((((..((....))..))))))).)))))))). (-23.05 = -22.83 + -0.22)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:31:26 2006