
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,236,271 – 12,236,406 |
| Length | 135 |
| Max. P | 0.980575 |

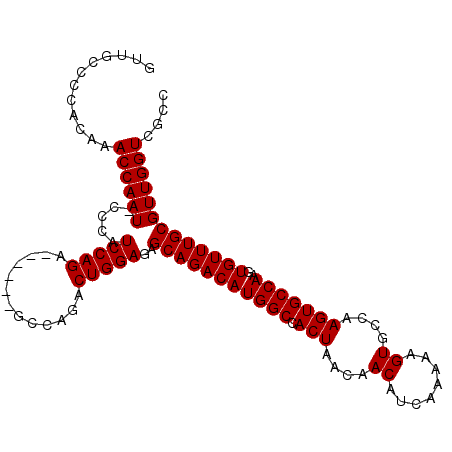
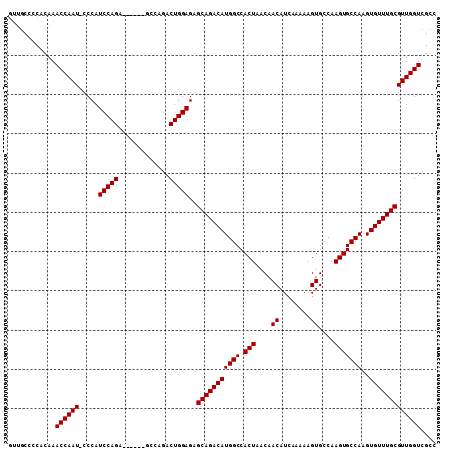
| Location | 12,236,271 – 12,236,373 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 90.62 |
| Mean single sequence MFE | -29.13 |
| Consensus MFE | -26.75 |
| Energy contribution | -26.75 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.803303 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12236271 102 + 22224390 GUUGCCCCACAAACCAAU-CCCAUCCAGA------GCCAGACUGGAGAGCAGACAUGGCCACUAACAACAUCAAAAAGUGCCAAGUGCCAAGUGUUUGCGUUGGUCGCC ............((((((-....(((((.------......)))))..(((((((((((.(((....((........))....)))))))..))))))))))))).... ( -27.50) >DroSim_CAF1 59953 102 + 1 GUUGCCCCACAAACCAAU-CCCAUCCAGA------ACCAGACUGGAGAGCAGACAUGGCCACUAACAACAUCAAAAAGUGCCAAGUGCCAAGUGUUUGCGUUGGUCGCC ............((((((-....(((((.------......)))))..(((((((((((.(((....((........))....)))))))..))))))))))))).... ( -27.50) >DroYak_CAF1 43903 105 + 1 ----CAACACAAACCAAUACCCAUCCAGAUGCAUGGGCAGACUGGAGAGCAGACAUGGCCACUAACAACAUCAAAAAGUGCCAAGUGCCAAGUGUUUGCGUUGGUCGCC ----........((((((.....(((((.(((....)))..)))))..(((((((((((.(((....((........))....)))))))..))))))))))))).... ( -32.40) >consensus GUUGCCCCACAAACCAAU_CCCAUCCAGA______GCCAGACUGGAGAGCAGACAUGGCCACUAACAACAUCAAAAAGUGCCAAGUGCCAAGUGUUUGCGUUGGUCGCC ............((((((.....(((((.............)))))..(((((((((((.(((....((........))....)))))))..))))))))))))).... (-26.75 = -26.75 + -0.00)

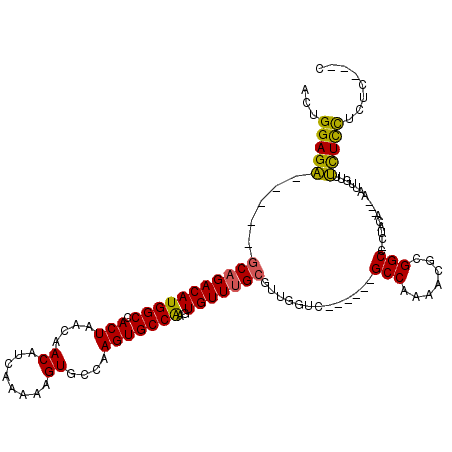
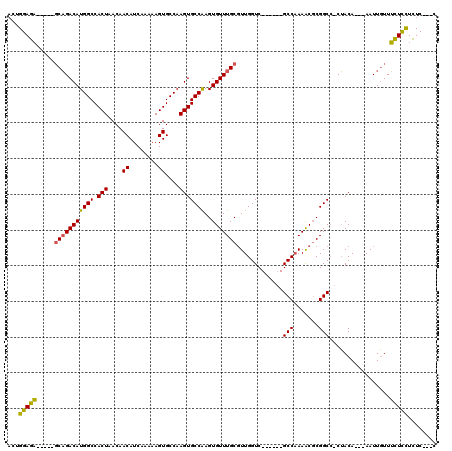
| Location | 12,236,304 – 12,236,406 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.33 |
| Mean single sequence MFE | -33.88 |
| Consensus MFE | -23.25 |
| Energy contribution | -22.62 |
| Covariance contribution | -0.62 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980575 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12236304 102 + 22224390 ACUGGAGA-----GCAGACAUGGCCACUAACAACAUCAAAAAGUGCCAAGUGCCAAGUGUUUGCGUUGGUC------GCCAAAACGCGGCC-CUACA---AAUUGUUUCUCCUCUC---C ...(((((-----(((((((((((.(((....((........))....)))))))..)))))))((.((((------((......))))))-..)).---.......)))))....---. ( -34.70) >DroVir_CAF1 29804 119 + 1 -CUGGAGCUGCACUCCGACAUGGCCACUAACAACAUCAAAAAGUGCCAAGUGCCGAGUGUUUGCCUUGGUCAGCGCUGCCCAAGCCCGGCACCGACAUUUAAUUUUUGUUUUUUUCCUCU -..((((.....))))((((((((.(((.............))))))).((((((((((((.((....)).))))))((....)).))))))..............)))).......... ( -30.52) >DroSim_CAF1 59986 102 + 1 ACUGGAGA-----GCAGACAUGGCCACUAACAACAUCAAAAAGUGCCAAGUGCCAAGUGUUUGCGUUGGUC------GCCAAAACGCGGCC-CUACA---AAUUGUUUCUCCUCUC---C ...(((((-----(((((((((((.(((....((........))....)))))))..)))))))((.((((------((......))))))-..)).---.......)))))....---. ( -34.70) >DroYak_CAF1 43939 105 + 1 ACUGGAGA-----GCAGACAUGGCCACUAACAACAUCAAAAAGUGCCAAGUGCCAAGUGUUUGCGUUGGUC------GCCAAAACGCGGCC-CUACA---AAUUGUUUCUCCUCUCCACC ..((((((-----(((((((((((.(((....((........))....)))))))..)))))))((.((((------((......))))))-..)).---............)))))).. ( -35.60) >consensus ACUGGAGA_____GCAGACAUGGCCACUAACAACAUCAAAAAGUGCCAAGUGCCAAGUGUUUGCGUUGGUC______GCCAAAACGCGGCC_CUACA___AAUUGUUUCUCCUCUC___C ...(((((.....(((((((((((.(((....((........))....)))))))..))))))).............(((.......))).................)))))........ (-23.25 = -22.62 + -0.62)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:31:18 2006