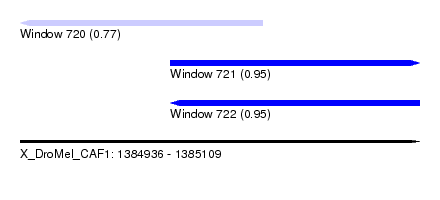
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,384,936 – 1,385,109 |
| Length | 173 |
| Max. P | 0.951271 |

| Location | 1,384,936 – 1,385,041 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.40 |
| Mean single sequence MFE | -37.90 |
| Consensus MFE | -27.28 |
| Energy contribution | -28.12 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.772453 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1384936 105 - 22224390 CACGUUGCUUGCUGCAUGCAGCAUGCUGCAUGAUGCAGCUGG------AGAUUAAAACACGACUUUGAUGCACUGAUGGCGUCUUUGAUUUUAGUCGCUGUC---------GCCUUUCAU .....(((..(((((((((((....)))))...))))))...------..((((((.......))))))))).(((.((((.(..((((....))))..).)---------)))..))). ( -31.10) >DroSec_CAF1 12346 114 - 1 CAUGUUGCUUGCUGCAUGCAGCAUGCUGCAUGCUGCAGCUGG------AGAUUAAAACACGACUUUGAUGCACUGAUGGCGUCUUUGAUUUUAGUCGCUGUCGUCGCUGUUGCCUUUCAU ......(((.((.((((((((....)))))))).)))))..(------(((....((((((((...(.....).(((((((.((........)).))))))))))).))))...)))).. ( -39.40) >DroSim_CAF1 12071 114 - 1 CAUGUUGCUUGCUGCAUGCAGCAUGCUGCAUGCUGCAGCUGG------AGAUUAAAACACGACUUUGAUGCACUGAUGGCGUCUUUGAUUUUAGUCGCUGUCGUCGCUGUUGCCUUUCAU ......(((.((.((((((((....)))))))).)))))..(------(((....((((((((...(.....).(((((((.((........)).))))))))))).))))...)))).. ( -39.40) >DroEre_CAF1 11313 111 - 1 CUGGCUGCAUGCUGCAUGCUGCAUGCCGCACGCUGCAGCUGG------AGAUUAAAACCCGACUUUGAUGCACAGAUGGCGUCUUUGAUUUUAGUCGCUGUC---GCCGUCGCCUUUCAU ..((((((((((((((.(((((.....))).))))))))(((------..........)))......)))))..(((((((.(..((((....))))..).)---)))))))))...... ( -41.60) >DroYak_CAF1 591 117 - 1 CAUGUUGAAUGUUGCAUGCUGCAUGCUGCAUGCUGCAGCUGGAGAUGGAGAUUAAAACCCGACUUUGAUGCACAGAUGGCGUCUUUGAUUUUAGUCGCUGUC---GCUGCUGCCUUUCAU .....((((....((((((........)))))).(((((.((.(((((.((((((((..(((....(((((.(....)))))).))).)))))))).)))))---.)))))))..)))). ( -38.00) >consensus CAUGUUGCUUGCUGCAUGCAGCAUGCUGCAUGCUGCAGCUGG______AGAUUAAAACACGACUUUGAUGCACUGAUGGCGUCUUUGAUUUUAGUCGCUGUC___GCUGUUGCCUUUCAU ...(((((.....((((((((....)))))))).)))))..........((((((((..(((....(((((.(....)))))).))).))))))))........................ (-27.28 = -28.12 + 0.84)

| Location | 1,385,001 – 1,385,109 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.08 |
| Mean single sequence MFE | -45.88 |
| Consensus MFE | -34.59 |
| Energy contribution | -35.19 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.947694 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1385001 108 + 22224390 AGCUGCAUCAUGCAGCAUGCUGCAUGCAGCAAGCAACGUGCAGCGGAGGCUCAACUGCGGAGCGGACAUCAGCG-----------UCCGAAUUUGGUGUGCUGAAUGCAGAAUCCAGA-A .(((((.....)))))...((((((.(((((.((....(((((..(.....)..)))))...(((((......)-----------))))......)).))))).))))))........-. ( -42.10) >DroSec_CAF1 12420 108 + 1 AGCUGCAGCAUGCAGCAUGCUGCAUGCAGCAAGCAACAUGCAGCGGAGGCUCAGCUGCGGAGCGGACAUCAGCG-----------UCCGAAUUUGGUGUGCUGAAUGCGGAAUCCAGA-A .((((((((((((((....)))))))).(....)....))))))(((.((((((((((.((.(((((......)-----------))))...)).))).)))))..))....)))...-. ( -46.50) >DroSim_CAF1 12145 108 + 1 AGCUGCAGCAUGCAGCAUGCUGCAUGCAGCAAGCAACAUGCAGCGGAGGCUCAGCUGCGGAGCGGACAUCAGCG-----------UCCGAAUUUGGUGUGCUGAAUGCGGAAUCCAGA-A .((((((((((((((....)))))))).(....)....))))))(((.((((((((((.((.(((((......)-----------))))...)).))).)))))..))....)))...-. ( -46.50) >DroEre_CAF1 11384 120 + 1 AGCUGCAGCGUGCGGCAUGCAGCAUGCAGCAUGCAGCCAGCAGCCGAGGCUCAACUGCGGAGCGGACAUCAGCGUGCCGCCUCCGUCCGAAUUUGGUGUGCUGAAUGCAGAGUCCCGAAA ..(((((((((((.(((((...))))).))))))...(((((((((((..((....(((((((((.(((....)))))).))))))..)).)))))).)))))..))))).......... ( -52.40) >DroYak_CAF1 668 119 + 1 AGCUGCAGCAUGCAGCAUGCAGCAUGCAACAUUCAACAUGCAGCGGAGGCUCAACUGCGGAGCGGACAUCAGCGUGCCGCCUCUGUCCGAAUUUGGUGUGCUGAAUGCAGGAUCCAGA-G ..((((((..((.(((.(((.(((((..........))))).)))...))))).)))))).((((.(((....)))))))((((((((..(((..(....)..)))...)))..))))-) ( -41.90) >consensus AGCUGCAGCAUGCAGCAUGCUGCAUGCAGCAAGCAACAUGCAGCGGAGGCUCAACUGCGGAGCGGACAUCAGCG___________UCCGAAUUUGGUGUGCUGAAUGCAGAAUCCAGA_A .((((((((((((((....)))))))).(....)....))))))...((.....(((((...((((((((((....................))))))).)))..)))))...))..... (-34.59 = -35.19 + 0.60)

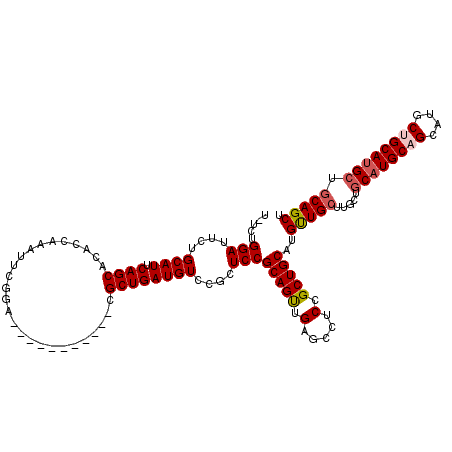
| Location | 1,385,001 – 1,385,109 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.08 |
| Mean single sequence MFE | -43.78 |
| Consensus MFE | -33.75 |
| Energy contribution | -34.35 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951271 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1385001 108 - 22224390 U-UCUGGAUUCUGCAUUCAGCACACCAAAUUCGGA-----------CGCUGAUGUCCGCUCCGCAGUUGAGCCUCCGCUGCACGUUGCUUGCUGCAUGCAGCAUGCUGCAUGAUGCAGCU .-.....((.((((((.(((((...(((...((((-----------((....))))))....(((((.(.....).)))))...)))..))))).)))))).))(((((.....))))). ( -38.50) >DroSec_CAF1 12420 108 - 1 U-UCUGGAUUCCGCAUUCAGCACACCAAAUUCGGA-----------CGCUGAUGUCCGCUCCGCAGCUGAGCCUCCGCUGCAUGUUGCUUGCUGCAUGCAGCAUGCUGCAUGCUGCAGCU .-...(((....((((.((((...((......)).-----------.))))))))....)))(((((.(.....).))))).....(((.((.((((((((....)))))))).))))). ( -42.90) >DroSim_CAF1 12145 108 - 1 U-UCUGGAUUCCGCAUUCAGCACACCAAAUUCGGA-----------CGCUGAUGUCCGCUCCGCAGCUGAGCCUCCGCUGCAUGUUGCUUGCUGCAUGCAGCAUGCUGCAUGCUGCAGCU .-...(((....((((.((((...((......)).-----------.))))))))....)))(((((.(.....).))))).....(((.((.((((((((....)))))))).))))). ( -42.90) >DroEre_CAF1 11384 120 - 1 UUUCGGGACUCUGCAUUCAGCACACCAAAUUCGGACGGAGGCGGCACGCUGAUGUCCGCUCCGCAGUUGAGCCUCGGCUGCUGGCUGCAUGCUGCAUGCUGCAUGCCGCACGCUGCAGCU ...........((((..((((....(((...((((((((.((((....))).).)))).))))...)))(((....)))))))..)))).((((((.(((((.....))).)))))))). ( -48.30) >DroYak_CAF1 668 119 - 1 C-UCUGGAUCCUGCAUUCAGCACACCAAAUUCGGACAGAGGCGGCACGCUGAUGUCCGCUCCGCAGUUGAGCCUCCGCUGCAUGUUGAAUGUUGCAUGCUGCAUGCUGCAUGCUGCAGCU .-..........((((((((((.........((((((..((((...))))..))))))....(((((.(.....).))))).)))))))))).....((((((.((.....)))))))). ( -46.30) >consensus U_UCUGGAUUCUGCAUUCAGCACACCAAAUUCGGA___________CGCUGAUGUCCGCUCCGCAGUUGAGCCUCCGCUGCAUGUUGCUUGCUGCAUGCAGCAUGCUGCAUGCUGCAGCU .....(((....((((.((((..........................))))))))....)))(((((.(.....).)))))..(((((.....((((((((....)))))))).))))). (-33.75 = -34.35 + 0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:45:20 2006