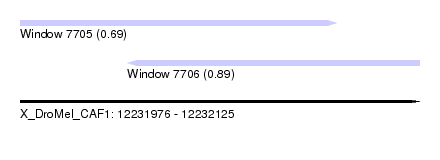
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,231,976 – 12,232,125 |
| Length | 149 |
| Max. P | 0.886991 |

| Location | 12,231,976 – 12,232,094 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 95.03 |
| Mean single sequence MFE | -26.96 |
| Consensus MFE | -23.44 |
| Energy contribution | -23.68 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.689843 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
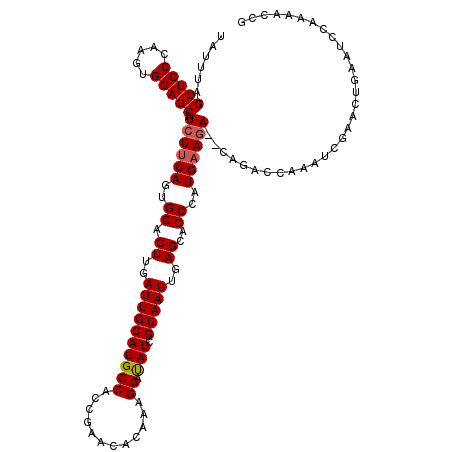
>X_DroMel_CAF1 12231976 118 + 22224390 UAUUUAGCUCCCAAGUGGAGCAUCUUCAGUGCACUUGAUUGCAUGCCACCAAACACAAAGGAUAUCGUAAUUGAGCAGCCAUGAAGACACAGACCAAAUCGAACUGAAUCCAAAACCG ......(((((.....))))).((((((..((.((..((((((((((............)).))).)))))..))..))..))))))..(((..(.....)..)))............ ( -26.20) >DroSec_CAF1 54494 116 + 1 UAUUUAGCUCCCAAGUGGAGCAUCUUCAGUGCUCUUGAUUGCAUGCCACCGAACACAAAGGAUAUCGUAAUUGAGCAGCCAUGAAGA--CAGACCAAAUCGAACUGAAUCCAAAACCG ......(((((.....))))).((((((..(((((..((((((((((............)).))).)))))..)).)))..))))))--(((..(.....)..)))............ ( -29.00) >DroSim_CAF1 55156 116 + 1 UAUUUAGCUCCCAAGUGGAGCAUCUUCAGUGCACUUGAUUGCAUGCCACCGAACACAAAGGAUAUCGUAAUUGAGCAGCCAUGAAGA--CAGACCAAAUCGAACUGAAUCCAAAACCG ......(((((.....))))).((((((..((.((..((((((((((............)).))).)))))..))..))..))))))--(((..(.....)..)))............ ( -26.20) >DroEre_CAF1 52167 116 + 1 UAUUUAGCUCCCAAGUGGAGCAUCUCCAUUGCACUUGAUUGCAUGCCACUAAACACAAAGGAUAUCGUAAUUGAGCAGCCAUGAAGA--CAGUCCAAAUCGAAGUGAAUCCAAAACCG (((((.(((((.....))))).(((.(((.((.((..((((((((((............)).))).)))))..))..)).))).)))--............)))))............ ( -23.30) >DroYak_CAF1 39594 116 + 1 UAUUUAGCUCCCAAGUGGAGCAUAUUCAUUGCACUUGAUUGCAUGCCACCGAACACAAAGGACAUCGUAAUUGAGCAGCCAUGAAGA--CAGUCCAAAUCGAGCUGCAUCCAAAACCG ....((((((.....((((.....(((((.((.((..((((((((((............)).))).)))))..))..)).)))))..--...))))....))))))............ ( -30.10) >consensus UAUUUAGCUCCCAAGUGGAGCAUCUUCAGUGCACUUGAUUGCAUGCCACCGAACACAAAGGAUAUCGUAAUUGAGCAGCCAUGAAGA__CAGACCAAAUCGAACUGAAUCCAAAACCG ......(((((.....))))).((((((..((.((..((((((((((............)).))).)))))..))..))..))))))............................... (-23.44 = -23.68 + 0.24)

| Location | 12,232,016 – 12,232,125 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 95.55 |
| Mean single sequence MFE | -34.44 |
| Consensus MFE | -29.50 |
| Energy contribution | -29.98 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886991 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
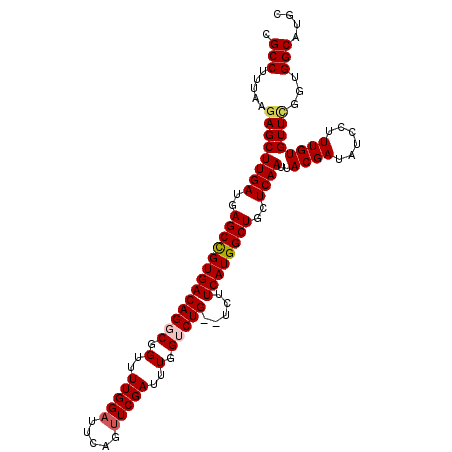
>X_DroMel_CAF1 12232016 109 - 22224390 CGCCUUUAAGAGCUUGAUGAGCCGUGACAGGCGGUUUUGGAUUCAGUUCGAUUUGGUCUGUGUCUUCAUGGCUGCUCAAUUACGAUAUCCUUUGUGUUUGGUGGCAUGC .(((.((((((((......(((((((((((((.(..((((((...))))))..).))))).....))))))))))))...(((((......))))).)))).))).... ( -33.60) >DroSec_CAF1 54534 107 - 1 CGCCUUUAAGAGCUUGAUGAGCCGUGACAGGCGGUUUUGGAUUCAGUUCGAUUUGGUCUG--UCUUCAUGGCUGCUCAAUUACGAUAUCCUUUGUGUUCGGUGGCAUGC .(((.....((((((((..((((((((.((((((..(..((((......))))..).)))--)))))))))))..))))..((((......))))))))...))).... ( -35.60) >DroSim_CAF1 55196 107 - 1 CGCCUUUAAGAGCUUGAUGAGCCGUGACAGGCGGUUUUGGAUUCAGUUCGAUUUGGUCUG--UCUUCAUGGCUGCUCAAUUACGAUAUCCUUUGUGUUCGGUGGCAUGC .(((.....((((((((..((((((((.((((((..(..((((......))))..).)))--)))))))))))..))))..((((......))))))))...))).... ( -35.60) >DroEre_CAF1 52207 107 - 1 CGCCUUUAAGAGCUUGAUGAGCUGUGACAGGCGGUUUUGGAUUCACUUCGAUUUGGACUG--UCUUCAUGGCUGCUCAAUUACGAUAUCCUUUGUGUUUAGUGGCAUGC .(((.((((((((......((((((((.(((((((.(..((((......))))..)))))--)))))))))))))))...(((((......))))).)))).))).... ( -34.00) >DroYak_CAF1 39634 107 - 1 CGCCUUUAAAAGCUUGAUGAGCCGUGACAGGCGGUUUUGGAUGCAGCUCGAUUUGGACUG--UCUUCAUGGCUGCUCAAUUACGAUGUCCUUUGUGUUCGGUGGCAUGC ((((.........((((..((((((((.(((((((.(..(((........)))..)))))--)))))))))))..)))).(((((......)))))...))))...... ( -33.40) >consensus CGCCUUUAAGAGCUUGAUGAGCCGUGACAGGCGGUUUUGGAUUCAGUUCGAUUUGGUCUG__UCUUCAUGGCUGCUCAAUUACGAUAUCCUUUGUGUUCGGUGGCAUGC .(((.....((((((((..(((((((((((((.(..(((((.....)))))..).))))).....))))))))..))))..((((......))))))))...))).... (-29.50 = -29.98 + 0.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:31:14 2006