| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,224,598 – 12,224,723 |
| Length | 125 |
| Max. P | 0.699334 |

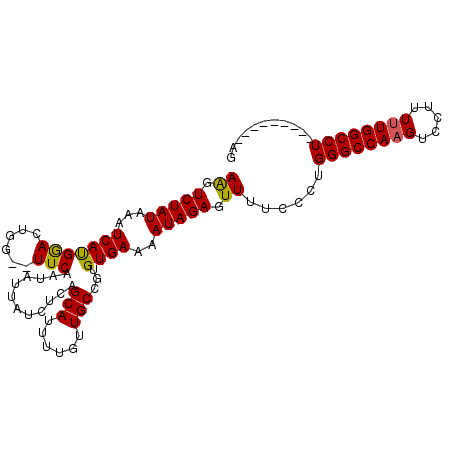
| Location | 12,224,598 – 12,224,689 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 90.92 |
| Mean single sequence MFE | -19.88 |
| Consensus MFE | -15.80 |
| Energy contribution | -16.30 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.548202 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12224598 91 + 22224390 UCAACGGGUAGCAAAACGAGACUAAAAGGACC---------AGGCCAAAAAGGACUUGGCCCAGGGAAAACUCUAUUUUCACACGGCAACAAAAUGCUGA ........(((((....((((......((.((---------(((((.....)).))))).))((((....))))..))))....(....)....))))). ( -21.70) >DroSec_CAF1 47420 90 + 1 UCAACGGGUAGCAAAACGAGACUAAAAAGACC---------AGGCCAAAAAGGACUUGGCCCAGGGA-AACUCUAUUUUCACACGGCAACAAAAUGCUGA .....(((((((.......).)))......((---------(((((.....)).))))))))(((..-..).)).........(((((......))))). ( -19.10) >DroEre_CAF1 44793 91 + 1 UCAACGGGUAGCAAAACGAGACUAAAAAGACU---------AGGCCAAAAAGGACAUGGCCCAGGGAAAACUCUAUUUUCACACGGCAACAAAAUGCUGC .......((((((....((((.........((---------.(((((.........))))).))(((....)))..))))....(....)....)))))) ( -19.10) >DroYak_CAF1 31970 100 + 1 UCAACGGGUAGCCAAACGAGACUAAAAAGACUAAAAAGUCCAGGCCAAAAUGGACUUGGCCCAGGGAAAACUCUAUUUUCACACCGCAACAAAAUGCUGA ......(((..((...............((((....))))..((((((.......))))))..))(((((.....)))))..)))(((......)))... ( -19.60) >consensus UCAACGGGUAGCAAAACGAGACUAAAAAGACC_________AGGCCAAAAAGGACUUGGCCCAGGGAAAACUCUAUUUUCACACGGCAACAAAAUGCUGA .................((((.....................((((((.......)))))).((((....))))..))))...(((((......))))). (-15.80 = -16.30 + 0.50)

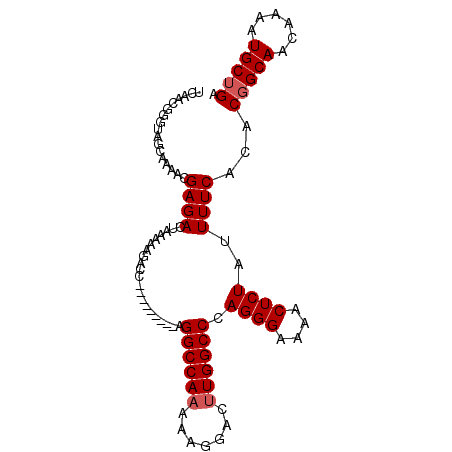

| Location | 12,224,628 – 12,224,723 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 87.06 |
| Mean single sequence MFE | -21.90 |
| Consensus MFE | -15.21 |
| Energy contribution | -16.02 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.627127 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12224628 95 + 22224390 CC---------AGGCCAAAAAGGACUUGGCCCAGGGAAAACUCUAUUUUCACACGGCAACAAAAUGCUGAGAUAAUACGGAA--CCAGUCCAUGAUUUAUAGACUU ((---------.((((((.......))))))...)).....(((((..(((..(((((......))))).........(((.--....))).)))...)))))... ( -23.20) >DroSec_CAF1 47450 94 + 1 CC---------AGGCCAAAAAGGACUUGGCCCAGGGA-AACUCUAUUUUCACACGGCAACAAAAUGCUGAGAUAAUACUGAA--CCAGUCCAUGAUUUAUAGACUU ((---------.((((((.......))))))...)).-...(((((..(((..(((((......))))).......((((..--.))))...)))...)))))... ( -21.30) >DroEre_CAF1 44823 95 + 1 CU---------AGGCCAAAAAGGACAUGGCCCAGGGAAAACUCUAUUUUCACACGGCAACAAAAUGCUGCGAUAAUAUGGAA--CCACUUCGUGAUUUAUAGACCU ((---------.(((((.........))))).)).......(((((..((((.(((((......)))))........((...--.))....))))...)))))... ( -21.60) >DroYak_CAF1 32000 106 + 1 CUAAAAAGUCCAGGCCAAAAUGGACUUGGCCCAGGGAAAACUCUAUUUUCACACCGCAACAAAAUGCUGAGAAAAUAUUGAACACUAGUCCAUGAUUUAUAGACUU .....(((((..((((((.......))))))((.(((....(((((((((.((..(((......))))).)))))))..)).......))).)).......))))) ( -21.50) >consensus CC_________AGGCCAAAAAGGACUUGGCCCAGGGAAAACUCUAUUUUCACACGGCAACAAAAUGCUGAGAUAAUACGGAA__CCAGUCCAUGAUUUAUAGACUU ............((((((.......))))))..........(((((..(((..(((((......))))).........(((.......))).)))...)))))... (-15.21 = -16.02 + 0.81)

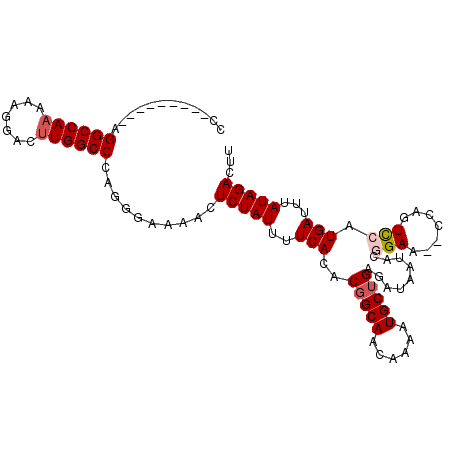

| Location | 12,224,628 – 12,224,723 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 87.06 |
| Mean single sequence MFE | -27.25 |
| Consensus MFE | -18.89 |
| Energy contribution | -18.57 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.699334 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12224628 95 - 22224390 AAGUCUAUAAAUCAUGGACUGG--UUCCGUAUUAUCUCAGCAUUUUGUUGCCGUGUGAAAAUAGAGUUUUCCCUGGGCCAAGUCCUUUUUGGCCU---------GG .(((((((.....)))))))((--..(..((((.((.(((((......)))..)).)).))))..)....))(..(((((((.....))))))).---------.) ( -25.30) >DroSec_CAF1 47450 94 - 1 AAGUCUAUAAAUCAUGGACUGG--UUCAGUAUUAUCUCAGCAUUUUGUUGCCGUGUGAAAAUAGAGUU-UCCCUGGGCCAAGUCCUUUUUGGCCU---------GG ((.(((((....(((((((((.--..)))).......((((.....))))))))).....))))).))-...(..(((((((.....))))))).---------.) ( -26.60) >DroEre_CAF1 44823 95 - 1 AGGUCUAUAAAUCACGAAGUGG--UUCCAUAUUAUCGCAGCAUUUUGUUGCCGUGUGAAAAUAGAGUUUUCCCUGGGCCAUGUCCUUUUUGGCCU---------AG ((.(((((....((((..(((.--...)))......(((((.....))))))))).....))))).))....((((((((.........))))))---------)) ( -27.60) >DroYak_CAF1 32000 106 - 1 AAGUCUAUAAAUCAUGGACUAGUGUUCAAUAUUUUCUCAGCAUUUUGUUGCGGUGUGAAAAUAGAGUUUUCCCUGGGCCAAGUCCAUUUUGGCCUGGACUUUUUAG ((((((.....((.(((((....))))).(((((((.(((((......)))..)).))))))))).........((((((((.....))))))))))))))..... ( -29.50) >consensus AAGUCUAUAAAUCAUGGACUGG__UUCAAUAUUAUCUCAGCAUUUUGUUGCCGUGUGAAAAUAGAGUUUUCCCUGGGCCAAGUCCUUUUUGGCCU_________AG ((.(((((...(((((((......)))............(((......)))...))))..))))).))......((((((((.....))))))))........... (-18.89 = -18.57 + -0.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:31:11 2006