| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,214,422 – 12,214,528 |
| Length | 106 |
| Max. P | 0.964866 |

| Location | 12,214,422 – 12,214,528 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 84.69 |
| Mean single sequence MFE | -32.00 |
| Consensus MFE | -23.52 |
| Energy contribution | -23.90 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959560 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
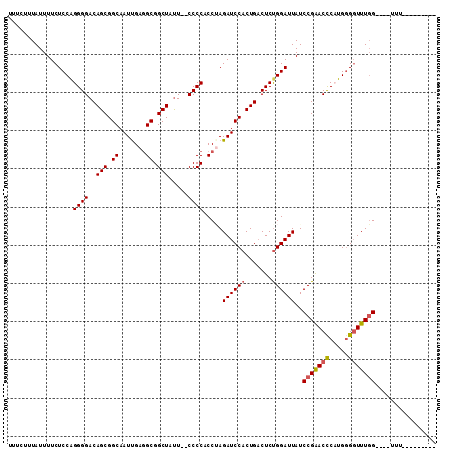
>X_DroMel_CAF1 12214422 106 + 22224390 UUUCUUUAUUUUCUCCAGGGGACAGCGGCAAUUGAGGCGGCUAUU--CCCCACCUAGAUCCACUGACUCUGGAUUAUCCGAACUCUUGGGGUUUGG----UUUUUUUUUUUU .............(((((((..(((.((..(((.(((.((.....--..)).))).))))).))).)))))))....(((((((.....)))))))----............ ( -29.90) >DroSec_CAF1 41144 97 + 1 UUUCUUUAUUUUCUCCAGGGGACAGCGGCAAUUGAUGCGGCUAUU--CCCCACCGAGAUCCACUGACUCCGGAUUAUCCGAACCCUUGAGGUUUGG----UUU--------- .............(((.(((((.(((.(((.....))).)))..)--))))...(((.((....))))).)))....(((((((.....)))))))----...--------- ( -31.00) >DroEre_CAF1 38657 101 + 1 UUUCUUUAUUUUCUCCGGGGGACAGCGGCAAUCGAGGCGGCUAUU--CCCCGCCUAGAUCCACUGCCUCUGGAUUAUCCGGACCCAUGGGGUUUGGUUUCUUU--------- .............((((((((.(((.((..(((.((((((.....--..)))))).))))).)))))))))))....((((((((...)))))))).......--------- ( -42.50) >DroYak_CAF1 26058 99 + 1 UUUCUUUAUUUUCUCCAGGGGACAGCGGCAAUUGAGGCGGCUACUCUCCCCACCAAGAUCCACUGACUCUGGAUUAUCCGAACCCAUGGGCUUUUG----CAU--------- ..............((((((((.(((.((.......)).)))....))))).....((((((.......))))))...........)))((....)----)..--------- ( -24.60) >consensus UUUCUUUAUUUUCUCCAGGGGACAGCGGCAAUUGAGGCGGCUAUU__CCCCACCUAGAUCCACUGACUCUGGAUUAUCCGAACCCAUGGGGUUUGG____UUU_________ .................((((..(((.((.......)).))).....)))).....((((((.......))))))..(((((((.....)))))))................ (-23.52 = -23.90 + 0.38)

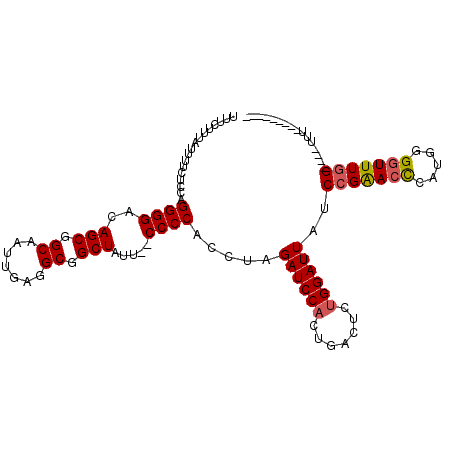
| Location | 12,214,422 – 12,214,528 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 84.69 |
| Mean single sequence MFE | -31.55 |
| Consensus MFE | -23.67 |
| Energy contribution | -25.18 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964866 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
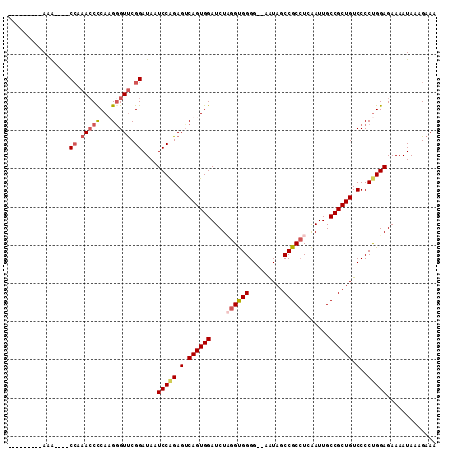
>X_DroMel_CAF1 12214422 106 - 22224390 AAAAAAAAAAAA----CCAAACCCCAAGAGUUCGGAUAAUCCAGAGUCAGUGGAUCUAGGUGGGG--AAUAGCCGCCUCAAUUGCCGCUGUCCCCUGGAGAAAAUAAAGAAA ............----((.(((.......))).))....(((((.(.((((((....((((((..--.....))))))......)))))).)..)))))............. ( -27.90) >DroSec_CAF1 41144 97 - 1 ---------AAA----CCAAACCUCAAGGGUUCGGAUAAUCCGGAGUCAGUGGAUCUCGGUGGGG--AAUAGCCGCAUCAAUUGCCGCUGUCCCCUGGAGAAAAUAAAGAAA ---------...----.....((....((.(((((.....))))).))...)).((((.(.((((--(.((((.(((.....))).)))))))))).))))........... ( -33.60) >DroEre_CAF1 38657 101 - 1 ---------AAAGAAACCAAACCCCAUGGGUCCGGAUAAUCCAGAGGCAGUGGAUCUAGGCGGGG--AAUAGCCGCCUCGAUUGCCGCUGUCCCCCGGAGAAAAUAAAGAAA ---------.......((..((((...))))..))....(((.(.(((((((((((.((((((..--.....)))))).)))..)))))).)).).)))............. ( -35.10) >DroYak_CAF1 26058 99 - 1 ---------AUG----CAAAAGCCCAUGGGUUCGGAUAAUCCAGAGUCAGUGGAUCUUGGUGGGGAGAGUAGCCGCCUCAAUUGCCGCUGUCCCCUGGAGAAAAUAAAGAAA ---------..(----(....))((((.(..((((.....)).))..).)))).((((.(.(((((.((..((..........))..)).)))))).))))........... ( -29.60) >consensus _________AAA____CCAAACCCCAAGGGUUCGGAUAAUCCAGAGUCAGUGGAUCUAGGUGGGG__AAUAGCCGCCUCAAUUGCCGCUGUCCCCUGGAGAAAAUAAAGAAA ................((.(((((...))))).))....(((((.(.((((((....((((((.........))))))......)))))).)..)))))............. (-23.67 = -25.18 + 1.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:31:05 2006