
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,214,196 – 12,214,287 |
| Length | 91 |
| Max. P | 0.998810 |

| Location | 12,214,196 – 12,214,287 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 72.67 |
| Mean single sequence MFE | -30.72 |
| Consensus MFE | -17.32 |
| Energy contribution | -19.10 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.26 |
| Mean z-score | -3.65 |
| Structure conservation index | 0.56 |
| SVM decision value | 3.23 |
| SVM RNA-class probability | 0.998810 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
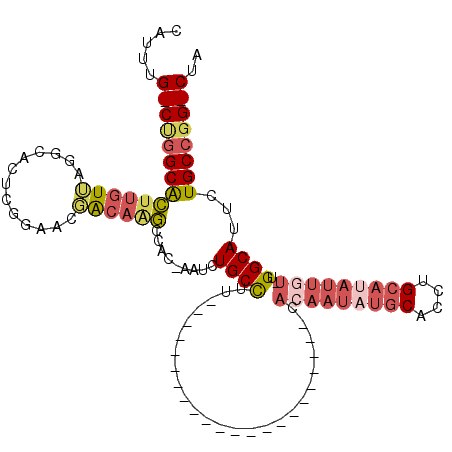
>X_DroMel_CAF1 12214196 91 + 22224390 CAUUUG-CUGGCACUUGUUAGGCACUCGGUACGACAAGCCAC-AAUCUGCCUU----------------------CACAAUAUGCACCUGCAUAUUGU-UGGCAUUCUGCCGG-CUA .....(-((((((((((((..((.....))..))))))....-....((((..----------------------.(((((((((....)))))))))-.))))...))))))-).. ( -35.20) >DroSec_CAF1 40918 91 + 1 CAUUUG-CUGGCACUUGUUAGGCACUCGGAACGACAAGCCAC-AAUCUGCCUU----------------------CACAAUAUGCACCUGCAUAUUGU-UGGCAUUCUGCCGG-CUA .....(-((((((.((((..(((..(((...)))...)))))-))..((((..----------------------.(((((((((....)))))))))-.))))...))))))-).. ( -34.00) >DroEre_CAF1 38413 91 + 1 CAUUUG-CUGGCACUUGUUAGGCACUCGGAACGACAAGCCAC-AAUCUGCCUU----------------------CACAAUAUGCACCUGCAUAUUGU-UGGCAUUCUGCCGG-CUA .....(-((((((.((((..(((..(((...)))...)))))-))..((((..----------------------.(((((((((....)))))))))-.))))...))))))-).. ( -34.00) >DroWil_CAF1 5041 77 + 1 CAUUUGGCUGGCAGCCCUCAAUCCCU-------GCCACCCGUACACCUGCCAC-------------------------------CACCUGGACAU-GU-UGGCAUGCUGCCUGUCUA .....(((.((((((..........(-------((((.......((.(((((.-------------------------------....))).)).-))-))))).)))))).))).. ( -20.91) >DroYak_CAF1 25813 91 + 1 CAUUUG-CUGGCACUUGUUAGGCACUCGGAACGACAAGCCGC-AAUCUGCCUU----------------------CACAAUAUGCACCUGCAUAUUGU-UGGCAUUCUGCCGG-CUA .....(-((((((.((((..(((..(((...)))...)))))-))..((((..----------------------.(((((((((....)))))))))-.))))...))))))-).. ( -34.30) >DroAna_CAF1 53086 110 + 1 CAUUUG-CCAGCGCUUGUAAGGA-C---AAAAUACAGGCCAC-AAUCUGCUUGCCACAAACACACAUACACCCCCCAGUAUAUGCACCUGCACAUUGUUUGGCAUUUUGCCGG-CUA .....(-((.(((((((((....-.---....)))))))...-........(((((..((((...((((........)))).(((....)))...)))))))))....)).))-).. ( -25.90) >consensus CAUUUG_CUGGCACUUGUUAGGCACUCGGAACGACAAGCCAC_AAUCUGCCUU______________________CACAAUAUGCACCUGCAUAUUGU_UGGCAUUCUGCCGG_CUA .....((((((((((((((.............)))))).........((((.........................(((((((((....)))))))))..))))...)))))))).. (-17.32 = -19.10 + 1.78)

| Location | 12,214,196 – 12,214,287 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 72.67 |
| Mean single sequence MFE | -32.69 |
| Consensus MFE | -16.63 |
| Energy contribution | -18.31 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.51 |
| SVM decision value | 2.44 |
| SVM RNA-class probability | 0.993951 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12214196 91 - 22224390 UAG-CCGGCAGAAUGCCA-ACAAUAUGCAGGUGCAUAUUGUG----------------------AAGGCAGAUU-GUGGCUUGUCGUACCGAGUGCCUAACAAGUGCCAG-CAAAUG ..(-(.(((....((((.-(((((((((....))))))))).----------------------..))))....-...((((((.((((...))))...))))))))).)-)..... ( -33.20) >DroSec_CAF1 40918 91 - 1 UAG-CCGGCAGAAUGCCA-ACAAUAUGCAGGUGCAUAUUGUG----------------------AAGGCAGAUU-GUGGCUUGUCGUUCCGAGUGCCUAACAAGUGCCAG-CAAAUG ..(-(.((((...((((.-(((((((((....))))))))).----------------------..))))..((-(((((...(((...)))..)))..)))).)))).)-)..... ( -33.10) >DroEre_CAF1 38413 91 - 1 UAG-CCGGCAGAAUGCCA-ACAAUAUGCAGGUGCAUAUUGUG----------------------AAGGCAGAUU-GUGGCUUGUCGUUCCGAGUGCCUAACAAGUGCCAG-CAAAUG ..(-(.((((...((((.-(((((((((....))))))))).----------------------..))))..((-(((((...(((...)))..)))..)))).)))).)-)..... ( -33.10) >DroWil_CAF1 5041 77 - 1 UAGACAGGCAGCAUGCCA-AC-AUGUCCAGGUG-------------------------------GUGGCAGGUGUACGGGUGGC-------AGGGAUUGAGGGCUGCCAGCCAAAUG .......(((.(.(((((-.(-((......)))-------------------------------.)))))).)))..((.((((-------((..........)))))).))..... ( -26.90) >DroYak_CAF1 25813 91 - 1 UAG-CCGGCAGAAUGCCA-ACAAUAUGCAGGUGCAUAUUGUG----------------------AAGGCAGAUU-GCGGCUUGUCGUUCCGAGUGCCUAACAAGUGCCAG-CAAAUG ..(-(.((((...((((.-(((((((((....))))))))).----------------------..))))....-..(((...(((...)))..))).......)))).)-)..... ( -32.10) >DroAna_CAF1 53086 110 - 1 UAG-CCGGCAAAAUGCCAAACAAUGUGCAGGUGCAUAUACUGGGGGGUGUAUGUGUGUUUGUGGCAAGCAGAUU-GUGGCCUGUAUUUU---G-UCCUUACAAGCGCUGG-CAAAUG ..(-(((((..........((((.(((((((((((((((((....))))))))).((((((...))))))....-...)))))))).))---)-)..........)))))-)..... ( -37.75) >consensus UAG_CCGGCAGAAUGCCA_ACAAUAUGCAGGUGCAUAUUGUG______________________AAGGCAGAUU_GUGGCUUGUCGUUCCGAGUGCCUAACAAGUGCCAG_CAAAUG ......(((.....)))..(((((((((....))))))))).........................((((.......(((..............))).......))))......... (-16.63 = -18.31 + 1.68)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:31:03 2006