| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,211,235 – 12,211,340 |
| Length | 105 |
| Max. P | 0.792776 |

| Location | 12,211,235 – 12,211,340 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 79.15 |
| Mean single sequence MFE | -38.15 |
| Consensus MFE | -17.89 |
| Energy contribution | -19.37 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.792776 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12211235 105 - 22224390 AACUCAGUGUCCUGCUGCACAAGGGCACUGCAGAGGAGACUACCUGGGAGUACACACUCUUCGUGGUCAACGACCACACCGCUUUCACGUGAGGCA----CAACAAAUU ..(((((((((((........))))))))).))((....))....((((((....))))...((((((...)))))).))(((((.....))))).----......... ( -35.60) >DroSec_CAF1 38022 98 - 1 ACCUCAGUGUCCUGCUCCACAAGGGCACUGCAGAGGAGACUACCUCGGAGUACACACUCUUCGUGGUCAACGACCACACCGCUUUCACGUGAGGCA----CA------- .((((((((((((........))))))))((.((((......))))(((((....)))))..((((((...))))))...))........))))..----..------- ( -36.80) >DroSim_CAF1 39484 98 - 1 ACCUCAGUGUCCUGCUCCACAAGGGCACUGCAGAGGAGACUACCUGGGAGUACACACUCUUCGUGGUCAACGACCACACCGCUUUCACGUGAGGCA----CA------- .((((((((((((........))))))))((..((....))....((((((....))))...((((((...)))))).))))........))))..----..------- ( -35.70) >DroEre_CAF1 35713 104 - 1 ACCUCAGUGUGCUGAUGCACAGGGGCACUGCCAAGGAGUCCUCCUGGGAGUAUACACUCUUCGUGGUCAACGACCACGCCGCCUACAGGUGAGGCA----CAACGAAU- .(((((.(((((....))))(((((((((.(((.(((....)))))).)))...........((((((...))))))))).)))...).)))))..----........- ( -41.70) >DroYak_CAF1 22381 105 - 1 AACUCAGUGUGCUGCUGCACAAGGGCACUGCGGAGGAGAUCACCUGGGAGUACACACUGCUGGUGGCCAACAACCACACCACUUUCAGGUGAGACA----CAACGAACU ....((((((.((........)).))))))((..(....(((((((..(((.((......))((((.......))))...)))..)))))))....----)..)).... ( -34.70) >DroPer_CAF1 3250 108 - 1 AUCUAAGUGUGCUCCUGUAUAGGGGCACUCCCGACGAGACUCCCUGGGAGUACACUCUGUUCGUGGUCAACGACGAGAAGGCCUUCAGGUGAGUGAGGCGUUCUCGCC- ..(..(((((((((((....(((((..(((.....)))..))))))))))))))))..).((((.....))))((((((.((((.((......)))))).))))))..- ( -44.40) >consensus ACCUCAGUGUCCUGCUGCACAAGGGCACUGCAGAGGAGACUACCUGGGAGUACACACUCUUCGUGGUCAACGACCACACCGCUUUCACGUGAGGCA____CAAC_AA__ ....((((((.((........)).))))))....((.........(((((......))))).((((((...)))))).))(((((.....))))).............. (-17.89 = -19.37 + 1.48)

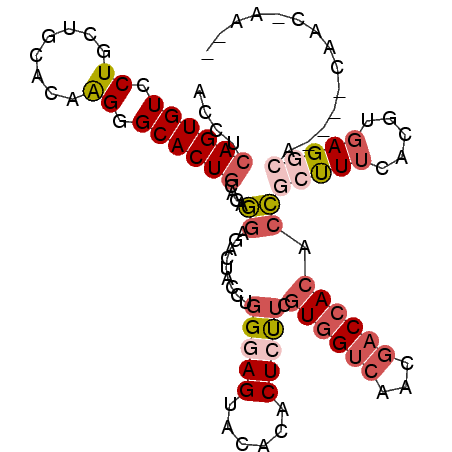

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:31:01 2006