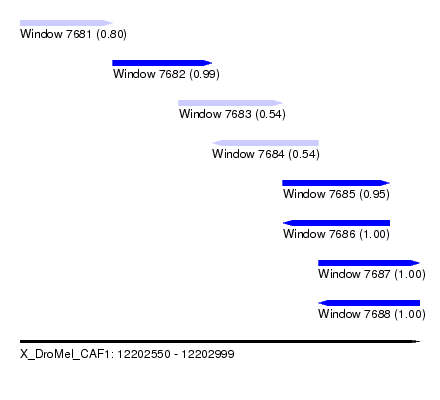
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,202,550 – 12,202,999 |
| Length | 449 |
| Max. P | 0.999831 |

| Location | 12,202,550 – 12,202,654 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 95.58 |
| Mean single sequence MFE | -24.38 |
| Consensus MFE | -22.38 |
| Energy contribution | -23.18 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.802613 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12202550 104 + 22224390 UUCCAAUUGCGUUGAUUUGUAAUAUUUGCUGUCGUUUCAUUAAUAUGCGAUAAGCAUUAUUGUUUCUGGGCAAAUGGCACAGCUCAUACACGAUUGCAAGACUG ...((((((.(((((.((((..(((((((((((((...........)))))(((((....)))))...))))))))..)))).))).)).))))))........ ( -25.60) >DroSec_CAF1 28839 104 + 1 UUCCAAUUGCGUUGAUUUGUAAUAUUUGCUGUCGUUUCAUUAAUAUGCGAUAAGCAUUAUUGUUUCUGGGCAAAUGGCACAGCUCAUACACGAUUGCAAGACUG ...((((((.(((((.((((..(((((((((((((...........)))))(((((....)))))...))))))))..)))).))).)).))))))........ ( -25.60) >DroSim_CAF1 30024 104 + 1 UUCCAAUUGCGUUGAUUUGUAAUAUUUGCUGUCGUUUCAUUAAUAUGCGAUAAGCAUUAUUGUUUCUGGGCAAAUGGCACAGCUCAUACACGAUUGCAAGACUG ...((((((.(((((.((((..(((((((((((((...........)))))(((((....)))))...))))))))..)))).))).)).))))))........ ( -25.60) >DroEre_CAF1 27960 104 + 1 UUCCAAUUGCGUUGAUUUGUUAUAUUUGCUGUCGUUUCAUUAAUAUGCGAUAAGCAUUAUUGUUUCUGGGCAAAUGGCACAGCUCAUACACGCUUGCAAGACUG ........(((((((.((((..(((((((((((((...........)))))(((((....)))))...))))))))..)))).)))...))))........... ( -22.40) >DroYak_CAF1 13767 98 + 1 UUCCAAUUGCGUUGAUUUGUUAUAUUUGCUGUCGUUUCAUUAAUAUGCGAUAAGCAUUAUUGUUUCUGGGCAAAUGGCACAGCGCAUACAAGACUG------UG .......(((((((...(((..(((((((((((((...........)))))(((((....)))))...))))))))))))))))))..........------.. ( -22.70) >consensus UUCCAAUUGCGUUGAUUUGUAAUAUUUGCUGUCGUUUCAUUAAUAUGCGAUAAGCAUUAUUGUUUCUGGGCAAAUGGCACAGCUCAUACACGAUUGCAAGACUG ...((((((.(((((.((((..(((((((((((((...........)))))(((((....)))))...))))))))..)))).))).)).))))))........ (-22.38 = -23.18 + 0.80)



| Location | 12,202,654 – 12,202,766 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 93.95 |
| Mean single sequence MFE | -28.40 |
| Consensus MFE | -24.12 |
| Energy contribution | -24.12 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.985547 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12202654 112 + 22224390 CAACACAUCGAUGAUUAUUGCGGCAUAAUUGUUGCAAAACUUGAACUCUUUGACCGUCCGGUCCAUCGGACAAUCGGCCAAGGAAUAAAGGACACUGUCCCCCAUUUCCAAC .......(((((.....((((((((....))))))))..................((((((....)))))).)))))....((((....((((...)))).....))))... ( -28.90) >DroSec_CAF1 28943 110 + 1 CAACACAUCGAUGAUUAUUGCGGUAUAAUUGUUGCAAAACUUGAACUCGUUGACCGUCCGGUCCAUCGGACAAUCGGCCAAGGAAUAAAGGACACUGUCC-CCAUUUCC-AC ........(((..(((((......)))))..)))..............(((((..((((((....))))))..)))))...((((....((((...))))-....))))-.. ( -26.20) >DroSim_CAF1 30128 110 + 1 CAACACAUCGAUGAUUAUUGCGGCAUAAUUGUUGCAAAACUUGAACUCUUUGACCGUCCGGUCCAUCGGACAAUCGGCCAAGGAAUAAAGGACACUGUCC-CCAUUUCC-AC .......(((((.....((((((((....))))))))..................((((((....)))))).)))))....((((....((((...))))-....))))-.. ( -29.50) >DroEre_CAF1 28064 110 + 1 CAACACAUCGAUGAUUAUUGCGGCAUAAUUGUUGCAAAACCUGAACUCUUUGACCGUCCGGUCCAUCGGACAAUCAGCCAAGGAAUAAAGGCUGCUGUCC-CCAUUUCC-AC .........((((....((((((((....))))))))..............((((....))))))))(((((..(((((..........))))).)))))-........-.. ( -32.50) >DroYak_CAF1 13865 106 + 1 CAACACAUCGAUGAUUAUUGCGGCAUAAUUGUUGCAAAACUUGAACUCUUUGAC----CAGUCCAUCGGACAAUCAGCCAAGGAAUAAAGGAUCCUGUCC-UCAUUUCC-AC ...........(((((.((((((((....)))))))).................----..((((...))))))))).....((((...(((((...))))-)...))))-.. ( -24.90) >consensus CAACACAUCGAUGAUUAUUGCGGCAUAAUUGUUGCAAAACUUGAACUCUUUGACCGUCCGGUCCAUCGGACAAUCGGCCAAGGAAUAAAGGACACUGUCC_CCAUUUCC_AC .......((((((....((((((((....))))))))..............((((....))))))))))............((((....((((...)))).....))))... (-24.12 = -24.12 + -0.00)


| Location | 12,202,728 – 12,202,845 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 95.97 |
| Mean single sequence MFE | -24.13 |
| Consensus MFE | -21.39 |
| Energy contribution | -21.17 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.535421 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12202728 117 + 22224390 CGGCCAAGGAAUAAAGGACACUGUCCCCCAUUUCCAACUCACAUCUACACAUCUGCGCAUCGCCGCACAGCCCGCUGGCCACCCUUCAUCCGGUCUUAGCCCCACUCCUGUGCCACA .(((((.((((....((((...)))).....)))).................(((.((......)).))).....)))))...........(((..(((........))).)))... ( -25.50) >DroSec_CAF1 29017 113 + 1 CGGCCAAGGAAUAAAGGACACUGUCC-CCAUUUCC-ACUCACAUCCACACAUCUGCGCAUCGCCGCACAACCCGCUGGCCACCCUUCAUCCGGUCUUAACCCCACUCCU--GCCACA .(((((.((((....((((...))))-....))))-.................((((......))))........)))))...........(((....)))........--...... ( -22.80) >DroSim_CAF1 30202 113 + 1 CGGCCAAGGAAUAAAGGACACUGUCC-CCAUUUCC-ACUCACAUCCACACAUCUGCGCAUCGCCGCACAGCCCGCUGGCCACCCUUCAUCCGGUCUUAACCCCACUCCU--GCCACA .(((((.((((....((((...))))-....))))-................(((.((......)).))).....)))))...........(((....)))........--...... ( -24.10) >consensus CGGCCAAGGAAUAAAGGACACUGUCC_CCAUUUCC_ACUCACAUCCACACAUCUGCGCAUCGCCGCACAGCCCGCUGGCCACCCUUCAUCCGGUCUUAACCCCACUCCU__GCCACA .(((((.((((....((((...)))).....))))..................((((......))))........)))))...........(((....)))................ (-21.39 = -21.17 + -0.22)


| Location | 12,202,766 – 12,202,885 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 97.18 |
| Mean single sequence MFE | -38.27 |
| Consensus MFE | -38.56 |
| Energy contribution | -37.90 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.32 |
| Structure conservation index | 1.01 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.535991 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12202766 119 - 22224390 UUUAUUAUGCAAAUCCGAGCCAGGGGUCGAAAACGAGGCGUGUGGCACAGGAGUGGGGCUAAGACCGGAUGAAGGGUGGCCAGCGGGCUGUGCGGCGAUGCGCAGAUGUGUAGAUGUGA ....(((((((..(((..((((.(.(((........))).).))))...)))((..(((((...((.......)).))))).))...(((((((....))))))).)))))))...... ( -40.60) >DroSec_CAF1 29053 117 - 1 UUUAUUAUGCAAAUCCGAGCCAGGGGUCGAAAACGAGGCGUGUGGC--AGGAGUGGGGUUAAGACCGGAUGAAGGGUGGCCAGCGGGUUGUGCGGCGAUGCGCAGAUGUGUGGAUGUGA ....(((((((..(((..((((.(.(((........))).).))))--.)))....(((((...((.......)).))))).(((.((((.....)))).)))...)))))))...... ( -36.30) >DroSim_CAF1 30238 117 - 1 UUUAUUAUGCAAAUCCGAGCCAGGGGUCGAAAACGAGGCGUGUGGC--AGGAGUGGGGUUAAGACCGGAUGAAGGGUGGCCAGCGGGCUGUGCGGCGAUGCGCAGAUGUGUGGAUGUGA ....(((((((..(((..((((.(.(((........))).).))))--.)))((..(((((...((.......)).))))).))...(((((((....))))))).)))))))...... ( -37.90) >consensus UUUAUUAUGCAAAUCCGAGCCAGGGGUCGAAAACGAGGCGUGUGGC__AGGAGUGGGGUUAAGACCGGAUGAAGGGUGGCCAGCGGGCUGUGCGGCGAUGCGCAGAUGUGUGGAUGUGA ....(((((((..(((..((((.(.(((........))).).))))...)))((..(((((...((.......)).))))).))...(((((((....))))))).)))))))...... (-38.56 = -37.90 + -0.66)

| Location | 12,202,845 – 12,202,965 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.22 |
| Mean single sequence MFE | -34.70 |
| Consensus MFE | -31.33 |
| Energy contribution | -31.83 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.949277 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12202845 120 + 22224390 CGCCUCGUUUUCGACCCCUGGCUCGGAUUUGCAUAAUAAAUGAGACACAAAAGCCAAGCGGCUACUGUCACAUGGCCAAUGGGGGAAACUGGCCAUGUGCCAUCUAUAAUGAUGAUAAUA .(.(((((((((((.(....).)))))...........)))))).)(((..((((....))))..)))((((((((((.....(....))))))))))).((((......))))...... ( -35.00) >DroSec_CAF1 29130 120 + 1 CGCCUCGUUUUCGACCCCUGGCUCGGAUUUGCAUAAUAAAUGAGACACAAAAGCCAAGCGGCUACUGUCACAUGGCCAAUGGGAGAAACUGGCCAUGUGCCAUCUAUAAUGAUGAUAAUA .(.(((((((((((.(....).)))))...........)))))).)(((..((((....))))..)))((((((((((...........)))))))))).((((......))))...... ( -33.60) >DroSim_CAF1 30315 120 + 1 CGCCUCGUUUUCGACCCCUGGCUCGGAUUUGCAUAAUAAAUGAGACACAAAAGCCAAGCGGCUACUGUCACAUGGCCAAUGGGGGAAACUGGCCAUGUGCCAUCUAUAAUGAUGAUAAUA .(.(((((((((((.(....).)))))...........)))))).)(((..((((....))))..)))((((((((((.....(....))))))))))).((((......))))...... ( -35.00) >DroEre_CAF1 28235 120 + 1 CGCCUCGCUUUCGACCCCUGGCUCGGAUUUGCAUAAUAAAUGAGACACAAAAGCCAAGCGGCUACUGUCACAUGGCCAAUGGGGGAAACUGGCCAUGUGCCAUCUAUAAUGAUGAUAAUA .((...((.(((((.(....).)))))...))...........((((....((((....))))..))))(((((((((.....(....))))))))))))((((......))))...... ( -35.40) >DroYak_CAF1 14030 120 + 1 CGCCUCGUUUUCGACCCCUGGCUCGGAUUUGCAUAAUAAAUGAGACACAAAAGCCAAGCGGCUACUGUCACAUGGCCAAUGGGGGAAACUGGCCAUGUGCCAUCUAUAAUGAUGAUAAUA .(.(((((((((((.(....).)))))...........)))))).)(((..((((....))))..)))((((((((((.....(....))))))))))).((((......))))...... ( -35.00) >DroAna_CAF1 42580 112 + 1 ------UCUGACGACCCCUGGCUCGGAUUUGCAUAAUAAAUGAGACACAA-AGCCAAGCGGCUACUGUCACAUGGCCAAUGG-GGAAACUGGCCAUGUGCCAUCUAUAAUGAUGAUAAUA ------(((((.(.((...))))))))...((...........((((...-((((....))))..))))(((((((((....-(....))))))))))))((((......))))...... ( -34.20) >consensus CGCCUCGUUUUCGACCCCUGGCUCGGAUUUGCAUAAUAAAUGAGACACAAAAGCCAAGCGGCUACUGUCACAUGGCCAAUGGGGGAAACUGGCCAUGUGCCAUCUAUAAUGAUGAUAAUA .........(((((.(....).)))))...((...........((((....((((....))))..))))(((((((((.....(....))))))))))))((((......))))...... (-31.33 = -31.83 + 0.50)



| Location | 12,202,845 – 12,202,965 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.22 |
| Mean single sequence MFE | -40.98 |
| Consensus MFE | -38.12 |
| Energy contribution | -38.12 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.93 |
| SVM decision value | 4.13 |
| SVM RNA-class probability | 0.999809 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12202845 120 - 22224390 UAUUAUCAUCAUUAUAGAUGGCACAUGGCCAGUUUCCCCCAUUGGCCAUGUGACAGUAGCCGCUUGGCUUUUGUGUCUCAUUUAUUAUGCAAAUCCGAGCCAGGGGUCGAAAACGAGGCG .....(((((......)))))((((((((((((.......))))))))))))......(((.((((((((((((((..........))))))....))))))))..(((....)))))). ( -41.30) >DroSec_CAF1 29130 120 - 1 UAUUAUCAUCAUUAUAGAUGGCACAUGGCCAGUUUCUCCCAUUGGCCAUGUGACAGUAGCCGCUUGGCUUUUGUGUCUCAUUUAUUAUGCAAAUCCGAGCCAGGGGUCGAAAACGAGGCG .....(((((......)))))((((((((((((.......))))))))))))......(((.((((((((((((((..........))))))....))))))))..(((....)))))). ( -41.30) >DroSim_CAF1 30315 120 - 1 UAUUAUCAUCAUUAUAGAUGGCACAUGGCCAGUUUCCCCCAUUGGCCAUGUGACAGUAGCCGCUUGGCUUUUGUGUCUCAUUUAUUAUGCAAAUCCGAGCCAGGGGUCGAAAACGAGGCG .....(((((......)))))((((((((((((.......))))))))))))......(((.((((((((((((((..........))))))....))))))))..(((....)))))). ( -41.30) >DroEre_CAF1 28235 120 - 1 UAUUAUCAUCAUUAUAGAUGGCACAUGGCCAGUUUCCCCCAUUGGCCAUGUGACAGUAGCCGCUUGGCUUUUGUGUCUCAUUUAUUAUGCAAAUCCGAGCCAGGGGUCGAAAGCGAGGCG .....(((((......)))))((((((((((((.......))))))))))))((((.((((....)))).))))(((((..............(((......)))(.(....))))))). ( -41.50) >DroYak_CAF1 14030 120 - 1 UAUUAUCAUCAUUAUAGAUGGCACAUGGCCAGUUUCCCCCAUUGGCCAUGUGACAGUAGCCGCUUGGCUUUUGUGUCUCAUUUAUUAUGCAAAUCCGAGCCAGGGGUCGAAAACGAGGCG .....(((((......)))))((((((((((((.......))))))))))))......(((.((((((((((((((..........))))))....))))))))..(((....)))))). ( -41.30) >DroAna_CAF1 42580 112 - 1 UAUUAUCAUCAUUAUAGAUGGCACAUGGCCAGUUUCC-CCAUUGGCCAUGUGACAGUAGCCGCUUGGCU-UUGUGUCUCAUUUAUUAUGCAAAUCCGAGCCAGGGGUCGUCAGA------ ................((((.((((((((((((....-..))))))))))))......(((.(((((((-(.(.....(((.....))).....).)))))))))))))))...------ ( -39.20) >consensus UAUUAUCAUCAUUAUAGAUGGCACAUGGCCAGUUUCCCCCAUUGGCCAUGUGACAGUAGCCGCUUGGCUUUUGUGUCUCAUUUAUUAUGCAAAUCCGAGCCAGGGGUCGAAAACGAGGCG .........(((.((((((((((((((((((((.......))))))))))))((((.((((....)))).))))...)))))))).)))......(((.(....).)))........... (-38.12 = -38.12 + 0.00)


| Location | 12,202,885 – 12,202,999 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.27 |
| Mean single sequence MFE | -31.78 |
| Consensus MFE | -27.72 |
| Energy contribution | -28.12 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.87 |
| SVM decision value | 3.30 |
| SVM RNA-class probability | 0.998952 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12202885 114 + 22224390 UGAGACACAAAAGCCAAGCGGCUACUGUCACAUGGCCAAUGGGGGAAACUGGCCAUGUGCCAUCUAUAAUGAUGAUAAUAAUGAUGCAAAUGCACACAC-UG-CACACACUGCAC----A ...((((....((((....))))..))))(((((((((.....(....))))))))))..((((......))))..........((((..((((.....-))-)).....)))).----. ( -32.90) >DroSec_CAF1 29170 105 + 1 UGAGACACAAAAGCCAAGCGGCUACUGUCACAUGGCCAAUGGGAGAAACUGGCCAUGUGCCAUCUAUAAUGAUGAUAAUAAUGAUGCAAAUGCACACAC-UG----------CAC----A ...((((....((((....))))..))))(((((((((...........)))))))))((((((......)))).......((.(((....)))))...-.)----------)..----. ( -30.30) >DroSim_CAF1 30355 105 + 1 UGAGACACAAAAGCCAAGCGGCUACUGUCACAUGGCCAAUGGGGGAAACUGGCCAUGUGCCAUCUAUAAUGAUGAUAAUAAUGAUGCAAAUGCACACAC-UG----------CAC----A ...((((....((((....))))..))))(((((((((.....(....))))))))))((((((......)))).......((.(((....)))))...-.)----------)..----. ( -31.70) >DroEre_CAF1 28275 99 + 1 UGAGACACAAAAGCCAAGCGGCUACUGUCACAUGGCCAAUGGGGGAAACUGGCCAUGUGCCAUCUAUAAUGAUGAUAAUAAUGAUGCAAAUGCACACAC--------------------- ...((((....((((....))))..))))(((((((((.....(....))))))))))..((((......)))).......((.(((....)))))...--------------------- ( -30.40) >DroAna_CAF1 42614 116 + 1 UGAGACACAA-AGCCAAGCGGCUACUGUCACAUGGCCAAUGG-GGAAACUGGCCAUGUGCCAUCUAUAAUGAUGAUAAUAAUGAUGCCAAAGGACAUCCACAUCG-ACA-AGGACAUGGG ...((((...-((((....))))..))))(((((((((....-(....)))))))))).((((((....(((((........((((((...)).))))..)))))-...-.)))..))). ( -33.60) >consensus UGAGACACAAAAGCCAAGCGGCUACUGUCACAUGGCCAAUGGGGGAAACUGGCCAUGUGCCAUCUAUAAUGAUGAUAAUAAUGAUGCAAAUGCACACAC_UG__________CAC____A ...((((....((((....))))..))))(((((((((.....(....))))))))))..((((......)))).......((.(((....)))))........................ (-27.72 = -28.12 + 0.40)


| Location | 12,202,885 – 12,202,999 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.27 |
| Mean single sequence MFE | -38.06 |
| Consensus MFE | -33.04 |
| Energy contribution | -33.08 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.78 |
| Structure conservation index | 0.87 |
| SVM decision value | 4.19 |
| SVM RNA-class probability | 0.999831 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12202885 114 - 22224390 U----GUGCAGUGUGUG-CA-GUGUGUGCAUUUGCAUCAUUAUUAUCAUCAUUAUAGAUGGCACAUGGCCAGUUUCCCCCAUUGGCCAUGUGACAGUAGCCGCUUGGCUUUUGUGUCUCA .----((((((.(((..-(.-....)..)))))))))........(((((......)))))((((((((((((.......))))))))))))((((.((((....)))).))))...... ( -41.50) >DroSec_CAF1 29170 105 - 1 U----GUG----------CA-GUGUGUGCAUUUGCAUCAUUAUUAUCAUCAUUAUAGAUGGCACAUGGCCAGUUUCUCCCAUUGGCCAUGUGACAGUAGCCGCUUGGCUUUUGUGUCUCA .----(((----------((-(((....)).))))))........(((((......)))))((((((((((((.......))))))))))))((((.((((....)))).))))...... ( -36.50) >DroSim_CAF1 30355 105 - 1 U----GUG----------CA-GUGUGUGCAUUUGCAUCAUUAUUAUCAUCAUUAUAGAUGGCACAUGGCCAGUUUCCCCCAUUGGCCAUGUGACAGUAGCCGCUUGGCUUUUGUGUCUCA .----(((----------((-(((....)).))))))........(((((......)))))((((((((((((.......))))))))))))((((.((((....)))).))))...... ( -36.50) >DroEre_CAF1 28275 99 - 1 ---------------------GUGUGUGCAUUUGCAUCAUUAUUAUCAUCAUUAUAGAUGGCACAUGGCCAGUUUCCCCCAUUGGCCAUGUGACAGUAGCCGCUUGGCUUUUGUGUCUCA ---------------------(((.((((....))))))).....(((((......)))))((((((((((((.......))))))))))))((((.((((....)))).))))...... ( -34.70) >DroAna_CAF1 42614 116 - 1 CCCAUGUCCU-UGU-CGAUGUGGAUGUCCUUUGGCAUCAUUAUUAUCAUCAUUAUAGAUGGCACAUGGCCAGUUUCC-CCAUUGGCCAUGUGACAGUAGCCGCUUGGCU-UUGUGUCUCA ..((((((..-...-.))))))((((((....)))))).......(((((......)))))((((((((((((....-..))))))))))))((((.((((....))))-))))...... ( -41.10) >consensus U____GUG__________CA_GUGUGUGCAUUUGCAUCAUUAUUAUCAUCAUUAUAGAUGGCACAUGGCCAGUUUCCCCCAUUGGCCAUGUGACAGUAGCCGCUUGGCUUUUGUGUCUCA .......................((((((....))).))).....(((((......)))))((((((((((((.......))))))))))))((((.((((....)))).))))...... (-33.04 = -33.08 + 0.04)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:30:58 2006