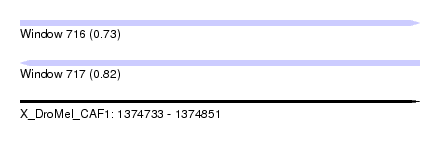
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,374,733 – 1,374,851 |
| Length | 118 |
| Max. P | 0.818008 |

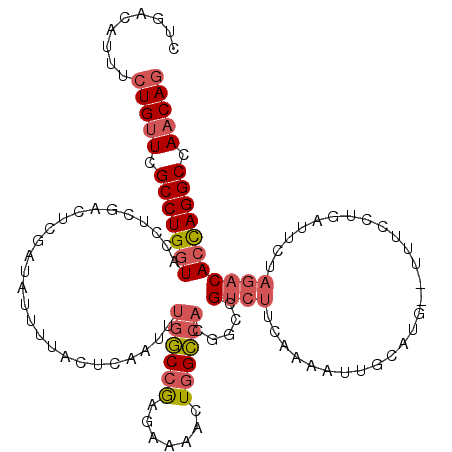
| Location | 1,374,733 – 1,374,851 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.85 |
| Mean single sequence MFE | -35.70 |
| Consensus MFE | -20.88 |
| Energy contribution | -22.82 |
| Covariance contribution | 1.94 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.729398 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1374733 118 + 22224390 CUGUUGGCCUGGUGUCUAGAAUAGAGAAA--CAUGCAAUUUUGAAGACGGCCGUGGCCAGUUUUCUCGGCCAAAUUGAGUAAAAUUUCGAGUCGUGGUACCAGGCGAACAGAAAUGCCAG (((((.((((((((.(((((...((((..--....((((((.(((((((((....))).))))))......)))))).......))))...)).))))))))))).)))))......... ( -41.42) >DroSec_CAF1 6769 118 + 1 CUGUUGGCCUAGUGUCUAGAAUCAGGAAA--CAUGCAAUUUUGAAGACUGCCGUGGCCAGUUUUCUCGGCCAAAUUGAGUAAAAUAUCCAGUCGAGGUACCAGGCGAACAGAAAUGUCAG (((((.((((.((..((.......(....--).............(((((...(((((((....)).)))))....((........))))))).))..)).)))).)))))......... ( -35.70) >DroSim_CAF1 7501 118 + 1 CUGUUGGCCUAGUGUCUAGAAUCAGGAAA--CAUGCAAUUUUGAAGACGGCCGUGGCCAGUUUUCUCGGCCAAAUUGAGUAAAAUAUCCAGUCGAGGUACCAGGCGAACAGAAAUGUCAG (((((.((((.((..((.......(....--).............((((((....))).((((((((((.....))))).))))).....))).))..)).)))).)))))......... ( -35.20) >DroEre_CAF1 5549 116 + 1 AUGAUCGCCUGGUGCAACAAUUAAUGAAAACUAUGAAAAUCGGA---CGUUUGAUCCGAGUUUUCUUGGGUAAAUUGAG-AGAUUCUGGGUUUCAGAUAACAGGCGAACAGAAAUGUCAG ....(((((((......(..(((((.....(((.((((((((((---.......))))).))))).)))....))))).-.)..((((.....))))...)))))))(((....)))... ( -30.50) >consensus CUGUUGGCCUAGUGUCUAGAAUAAGGAAA__CAUGCAAUUUUGAAGACGGCCGUGGCCAGUUUUCUCGGCCAAAUUGAGUAAAAUAUCCAGUCGAGGUACCAGGCGAACAGAAAUGUCAG (((((.(((((((((((.((....(((.......((.............))..(((((((....)).)))))..............)))..)).))))))))))).)))))......... (-20.88 = -22.82 + 1.94)

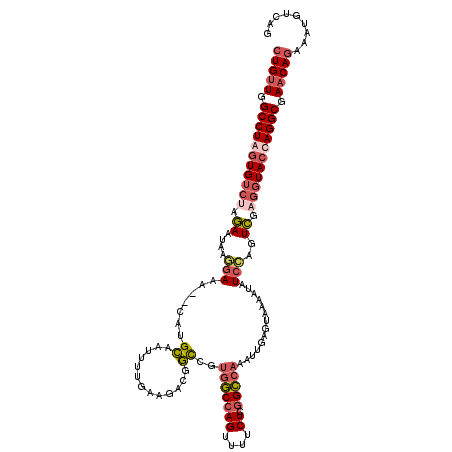
| Location | 1,374,733 – 1,374,851 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.85 |
| Mean single sequence MFE | -30.70 |
| Consensus MFE | -18.79 |
| Energy contribution | -20.23 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.818008 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1374733 118 - 22224390 CUGGCAUUUCUGUUCGCCUGGUACCACGACUCGAAAUUUUACUCAAUUUGGCCGAGAAAACUGGCCACGGCCGUCUUCAAAAUUGCAUG--UUUCUCUAUUCUAGACACCAGGCCAACAG .........(((((.(((((((..........(((((......((((((....((((....((((....))))))))..))))))...)--))))((((...)))).))))))).))))) ( -30.70) >DroSec_CAF1 6769 118 - 1 CUGACAUUUCUGUUCGCCUGGUACCUCGACUGGAUAUUUUACUCAAUUUGGCCGAGAAAACUGGCCACGGCAGUCUUCAAAAUUGCAUG--UUUCCUGAUUCUAGACACUAGGCCAACAG .........(((((.(((((((..((.((..(((..............((((((.......))))))..(((((.......)))))...--..)))....)).))..))))))).))))) ( -32.20) >DroSim_CAF1 7501 118 - 1 CUGACAUUUCUGUUCGCCUGGUACCUCGACUGGAUAUUUUACUCAAUUUGGCCGAGAAAACUGGCCACGGCCGUCUUCAAAAUUGCAUG--UUUCCUGAUUCUAGACACUAGGCCAACAG .........(((((.(((((((..((((.(..(((..........)))..).))))......(((....)))(((((((((((.....)--)))..)))....))))))))))).))))) ( -32.60) >DroEre_CAF1 5549 116 - 1 CUGACAUUUCUGUUCGCCUGUUAUCUGAAACCCAGAAUCU-CUCAAUUUACCCAAGAAAACUCGGAUCAAACG---UCCGAUUUUCAUAGUUUUCAUUAAUUGUUGCACCAGGCGAUCAU ..........((.(((((((...((((.....))))....-..............(((((.((((((.....)---))))))))))((((((......)))))).....))))))).)). ( -27.30) >consensus CUGACAUUUCUGUUCGCCUGGUACCUCGACUCGAUAUUUUACUCAAUUUGGCCGAGAAAACUGGCCACGGCCGUCUUCAAAAUUGCAUG__UUUCCUGAUUCUAGACACCAGGCCAACAG .........(((((.(((((((..........................((((((.......)))))).....((((...........................))))))))))).))))) (-18.79 = -20.23 + 1.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:45:16 2006