
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,195,072 – 12,195,256 |
| Length | 184 |
| Max. P | 0.934791 |

| Location | 12,195,072 – 12,195,184 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.42 |
| Mean single sequence MFE | -35.03 |
| Consensus MFE | -15.76 |
| Energy contribution | -18.07 |
| Covariance contribution | 2.31 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.45 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.934791 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12195072 112 - 22224390 GGGCGGGUAGGUGCGGCAG---GUUGGCUGCCGUGCAAAAGGUAAACACAAAUUGUUGCAUGGCAGCGCUGAAGCGAUGGCUUACAGUGCGGCAAAAAUCG-----UUGGUAAUAGAAUG .(((((.....(((.((..---....((((((((((((...((....))......))))))))))))((((((((....)))).)))))).)))....)))-----))............ ( -42.80) >DroSec_CAF1 21377 112 - 1 GGGCGGCUGGGUGCGGCAG---GUUGGCUGCCGUGCAAAAGGUAAACACAAAUUGUUGCAUGGAAGCGCUGAAGCGAUGGCUUACAGUGCGGCAAAAAUCG-----UUGUUAAUAAAAAG .((((((...(..((((((---.....))))))..)...................((((......((((((((((....)))).)))))).)))).....)-----)))))......... ( -37.50) >DroSim_CAF1 22601 112 - 1 GGGCGGGUGGGUGCGGCAG---GUUGGCUGCCGUGCAAAAGGUAAACACAAAUUGUUGCAUGGAAGCGCUGAAGCGAUGGCUUACAGUGCGGCAAAAAUCG-----UUGGUAAUAAAAAG .((((((((.(..((((((---.....))))))..)..........)))......((((......((((((((((....)))).)))))).))))...)))-----))............ ( -37.00) >DroEre_CAF1 20767 95 - 1 GAGGCGGUGGGAGUGGCAG---GUUGGCUGCCGUGCAAAAGGUAAACACAAAUUGUUGCAU-----------------GGCGUACAGUGCGGCAAAAAUCU-----GUGGUUACAAAGAG ....(..((..(.(.((((---(((.((((((((((((...((....))......))))))-----------------)))((.....)))))...)))))-----)).))..))..).. ( -26.80) >DroYak_CAF1 5952 112 - 1 CUGAGGGCGGGUGUGGCAG---GUUGGCUGCCGUGCAAAAGGUAAACACAAAUUGUUGCAUGGGAGCGCUCAAGCGAUGGCUUACAGUGCGCCAAAAAUCC-----GUGAUAAUAAAAUG ......((((((.((((..---....(((.((((((((...((....))......)))))))).)))((.(((((....))))...).))))))...))))-----))............ ( -37.70) >DroAna_CAF1 33355 118 - 1 CGGCAGCCAGAUCCUGGAGCAUAGGGAGUGGCAUCCACAAGGUAAACACAAAUUGUUGCAUGGGUGCAGUGAUGCGGAGAUGGGCACUGAA--AGAAAUUCUAAAGGUGUUAUUAAGAUA (.((.((((..(((((.....)))))..))))...................((..(((((....)))))..)))).).....((((((...--((.....))...))))))......... ( -28.40) >consensus GGGCGGGUGGGUGCGGCAG___GUUGGCUGCCGUGCAAAAGGUAAACACAAAUUGUUGCAUGGAAGCGCUGAAGCGAUGGCUUACAGUGCGGCAAAAAUCG_____GUGGUAAUAAAAAG ..........(((((((((.....((..((((........))))..))....)))))))))....((((((((((....)))).)))))).............................. (-15.76 = -18.07 + 2.31)

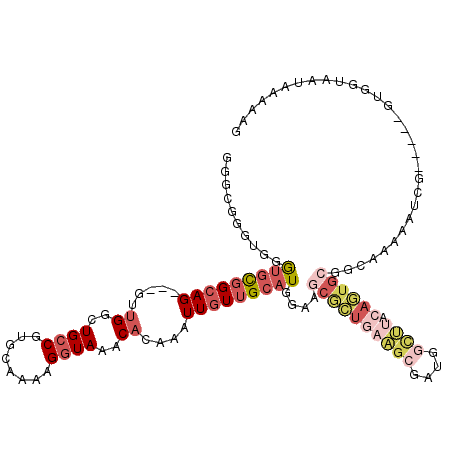

| Location | 12,195,147 – 12,195,256 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 84.85 |
| Mean single sequence MFE | -37.16 |
| Consensus MFE | -23.94 |
| Energy contribution | -25.06 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.705955 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12195147 109 - 22224390 AAUGAAGCUCGAAAUGAAAAGCCAGCCCAUGCAAAAAACGGCAAGGGGGGCGUGGUCG---U-----GGGUGAAUGUGAGGGGCGGGUAGGUGCGGCAGGUUGGCUGCCGUGCAAAA ......(((((..((((...(((..(((.(((........))).))).)))....)))---)-----................)))))..(..((((((.....))))))..).... ( -36.77) >DroSec_CAF1 21452 107 - 1 AAUGAAGCUCGAAAUGAAAAGCCAGCCCAUGCAAAAACCGGCAAGG-GGGCGUGGUCG---U-----GGGUGAAUGUG-GGGGCGGCUGGGUGCGGCAGGUUGGCUGCCGUGCAAAA ...................((((.((((..(((...(((.((....-..........)---)-----.)))...))).-.))))))))..(..((((((.....))))))..).... ( -38.24) >DroSim_CAF1 22676 107 - 1 AAUGAAGCUCGAAAUGAAAAGCCAGCCCAUGCAAAAAACGGCAAGG-GGGCGUGGUCG---U-----GGGCGAAUGUG-GGGGCGGGUGGGUGCGGCAGGUUGGCUGCCGUGCAAAA ......((((....(....)(((.((((((((.....(((.(....-.).))).).))---)-----)))).......-..)))))))..(..((((((.....))))))..).... ( -37.40) >DroEre_CAF1 20825 106 - 1 AAUGAAGCUCGAAAUGAAAAGCCAGCCAGUGCAAAAAACAGCAAGG-G-GCGUGGCUG---U-----GGGCGGGUGUG-GGAGGCGGUGGGAGUGGCAGGUUGGCUGCCGUGCAAAA ......(((((........((((((((..(((........)))...-)-)).))))).---)-----))))..(..((-(.((.(..(..(.....)..)..).)).)))..).... ( -34.20) >DroYak_CAF1 6027 114 - 1 AAUGAAGCUCGAAAUGAAAAGCCAGCCUGUGCAAAAAACAGCCAGG-C-GGGUGGGCGGUGUUGGUUAAGGGGGCGUG-GCUGAGGGCGGGUGUGGCAGGUUGGCUGCCGUGCAAAA ......((((..........(((.(((((.((........))))))-)-.))).(((.(((((.........))))).-)))..))))..(..((((((.....))))))..).... ( -39.20) >consensus AAUGAAGCUCGAAAUGAAAAGCCAGCCCAUGCAAAAAACGGCAAGG_GGGCGUGGUCG___U_____GGGCGAAUGUG_GGGGCGGGUGGGUGCGGCAGGUUGGCUGCCGUGCAAAA ......((((..........((((((((.(((........)))....)))).))))...........))))...................(..((((((.....))))))..).... (-23.94 = -25.06 + 1.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:30:45 2006