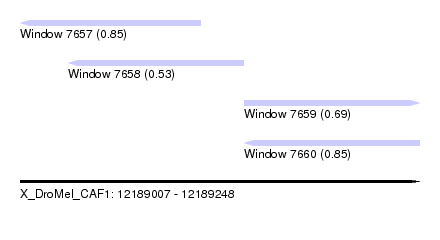
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,189,007 – 12,189,248 |
| Length | 241 |
| Max. P | 0.854469 |

| Location | 12,189,007 – 12,189,116 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.14 |
| Mean single sequence MFE | -32.33 |
| Consensus MFE | -23.83 |
| Energy contribution | -23.70 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.853817 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12189007 109 - 22224390 CUUAAAAGUCGAUGCGCAAGCAAGGCUAAAACACUUUUGCAGUCACUCAAGUGACAAUGCAGACGGCCAGUGGGAAAUUGGGGGCAGGGAAAAGUGGGU----GGGAGC-------GAUG .......(((..(((....)))..(((.(..(((((((.(.(((((....)))))..(((......(((((.....)))))..))).).)))))))..)----...)))-------))). ( -34.00) >DroSec_CAF1 15339 107 - 1 CUUAAAAGUCGAUGCGCAAGCAAGGCUAAAACACUUUUGCAGUCACUCAAGUGACAAUUCAGACGGCCAGUGGGAAAUUGGGGGCAGGGAAAAGUGGGC----GG--GC-------GAUG .......(((..(((....)))..(((....(((((((.(.(((((....)))))...........(((((.....)))))......).)))))))...----.)--))-------))). ( -32.60) >DroEre_CAF1 15278 93 - 1 CUUAAAAGUCGAUGCGCAAGCAAGGCUAAAACACUUUUGCAGUCACUCAAGUGACAAUGCAGACGGCCAGUGG--------------GGGAAAGUGGGU----GG--GU-------GAUG ......((((..(((....))).))))....(((((((((((((((....)))))..))))))((.(((.(..--------------.....).))).)----))--))-------)... ( -30.20) >DroAna_CAF1 28050 117 - 1 CUUAAAAGUCGAUGUGCAAGCAAGGCUAAAACACUUUUGCAGUCACUCAAGUGACAAUGCAGACGGCCACUGGGAAAAUG---UCCUGGAAAAGUGGGAAAGUGGGCGCCACUAGUGAUG ......((((..(((....))).))))....(((((((((((((((....)))))..))))))((.(((((.........---((((........)))).))))).)).....))))... ( -32.50) >consensus CUUAAAAGUCGAUGCGCAAGCAAGGCUAAAACACUUUUGCAGUCACUCAAGUGACAAUGCAGACGGCCAGUGGGAAAUUG___GCAGGGAAAAGUGGGU____GG__GC_______GAUG ......((((..(((....))).))))....(((((((.(.(((((....)))))...........((....)).............).)))))))........................ (-23.83 = -23.70 + -0.13)



| Location | 12,189,036 – 12,189,142 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.22 |
| Mean single sequence MFE | -34.02 |
| Consensus MFE | -22.83 |
| Energy contribution | -22.70 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.67 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.529805 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12189036 106 - 22224390 GGGUUUC-----------G--GCGGCGAUGGG-GCAACUACUUAAAAGUCGAUGCGCAAGCAAGGCUAAAACACUUUUGCAGUCACUCAAGUGACAAUGCAGACGGCCAGUGGGAAAUUG ...((((-----------.--(((((....((-....)).......((((..(((....))).))))......(.(((((((((((....)))))..)))))).)))).)).)))).... ( -33.50) >DroSec_CAF1 15366 106 - 1 GGGUUUC-----------G--GCGGGGAUGGG-GCAACUACUUAAAAGUCGAUGCGCAAGCAAGGCUAAAACACUUUUGCAGUCACUCAAGUGACAAUUCAGACGGCCAGUGGGAAAUUG .((((((-----------(--((.(...((((-((((.........((((..(((....))).)))).........)))).(((((....)))))..))))..).))).....)))))). ( -29.97) >DroEre_CAF1 15298 99 - 1 GGGUUGC-----------G--CCGGGAAUGGG-GCAACUACUUAAAAGUCGAUGCGCAAGCAAGGCUAAAACACUUUUGCAGUCACUCAAGUGACAAUGCAGACGGCCAGUGG------- .((((((-----------.--(((....))).-)))))).......((((..(((....))).))))....(((((((((((((((....)))))..)))))).....)))).------- ( -37.30) >DroAna_CAF1 28087 120 - 1 AGGCGGCCAGGAAGGAGGGCUGGGGAGCUGGGGCCAACUACUUAAAAGUCGAUGUGCAAGCAAGGCUAAAACACUUUUGCAGUCACUCAAGUGACAAUGCAGACGGCCACUGGGAAAAUG .((.(((((((.((..((.((.((...)).)).))..)).)))....(((....((((.((((((.........)))))).(((((....)))))..))))))))))).))......... ( -35.30) >consensus GGGUUGC___________G__GCGGGGAUGGG_GCAACUACUUAAAAGUCGAUGCGCAAGCAAGGCUAAAACACUUUUGCAGUCACUCAAGUGACAAUGCAGACGGCCAGUGGGAAAUUG .(((..........................................((((..(((....))).))))......(.(((((((((((....)))))..)))))).))))............ (-22.83 = -22.70 + -0.13)

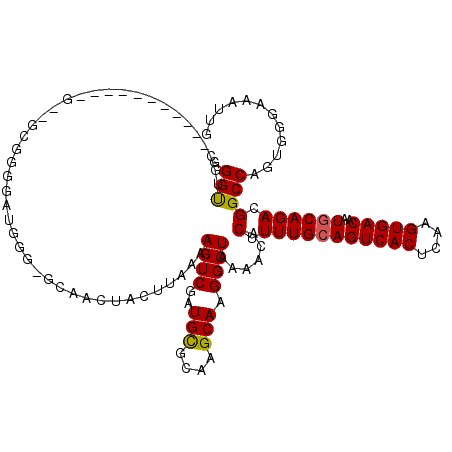
| Location | 12,189,142 – 12,189,248 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 87.88 |
| Mean single sequence MFE | -43.10 |
| Consensus MFE | -33.42 |
| Energy contribution | -34.87 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.691584 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12189142 106 + 22224390 GAGCACUUUCCCCGCCCACUUUCCAUUCCCCAUUUUCCAUUUCCCUUGCCAAAUGCUAGUGCAUCGGCGGU----GCAAGGUGGUGGGGGCAAAAGCGAGGGAGUUGCUC (((((..((((((((....(((....((((((....((((((..((((((..((((....)))).))))).----)..))))))))))))..)))))).))))).))))) ( -38.10) >DroSec_CAF1 15472 109 + 1 GAGCACUUUCCCCGCCCACUUUCCAUUCCCCAUUUUCCAUUUUCCCAGCCAAAUGCAAGUGCAUCGGCGGUGGGUGCGAGG-AGUGGGGGCAAAUGCGAGGGAGUUGCUC (((((..((((((((....(..(((((((((((...(((.....((.(((..((((....)))).))))))))))).).))-)))))..).....))).))))).))))) ( -43.10) >DroEre_CAF1 15397 107 + 1 GAGCACUUUCCCCGCCCACUUUCCAUUGCCCAUUUUCCAUUUC-CCAGCCAAAUGCAAGUGCAUCGGCGGAGGGAGC-A-GUGGUGGGGGAAAGUGCGAGGGAGCUGCUC (((((..((((((((..(((((((....((((....(((((((-((.(((..((((....)))).)))...))))..-)-)))))))))))))))))).))))).))))) ( -48.10) >consensus GAGCACUUUCCCCGCCCACUUUCCAUUCCCCAUUUUCCAUUUCCCCAGCCAAAUGCAAGUGCAUCGGCGGUGGG_GC_AGGUGGUGGGGGCAAAUGCGAGGGAGUUGCUC (((((..((((((((....(..((((..((.....((((.....((.(((..((((....)))).))))))))).....))..))))..).....))).))))).))))) (-33.42 = -34.87 + 1.45)



| Location | 12,189,142 – 12,189,248 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 87.88 |
| Mean single sequence MFE | -45.53 |
| Consensus MFE | -32.89 |
| Energy contribution | -33.00 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.854469 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12189142 106 - 22224390 GAGCAACUCCCUCGCUUUUGCCCCCACCACCUUGC----ACCGCCGAUGCACUAGCAUUUGGCAAGGGAAAUGGAAAAUGGGGAAUGGAAAGUGGGCGGGGAAAGUGCUC (((((.(((((((((((((..((((((((((((((----......(((((....)))))..))))))....)))....)))))....))))))))).))))....))))) ( -43.40) >DroSec_CAF1 15472 109 - 1 GAGCAACUCCCUCGCAUUUGCCCCCACU-CCUCGCACCCACCGCCGAUGCACUUGCAUUUGGCUGGGAAAAUGGAAAAUGGGGAAUGGAAAGUGGGCGGGGAAAGUGCUC (((((..((((.(((.(((.((((((.(-((.....((((..((((((((....))).))))))))).....)))...)))))...).)))....)))))))...))))) ( -45.10) >DroEre_CAF1 15397 107 - 1 GAGCAGCUCCCUCGCACUUUCCCCCACCAC-U-GCUCCCUCCGCCGAUGCACUUGCAUUUGGCUGG-GAAAUGGAAAAUGGGCAAUGGAAAGUGGGCGGGGAAAGUGCUC (((((..((((((.(((((((((((((((.-.-..((((...((((((((....))).))))).))-))..)))....))))....)))))))).).)))))...))))) ( -48.10) >consensus GAGCAACUCCCUCGCAUUUGCCCCCACCACCU_GC_CCCACCGCCGAUGCACUUGCAUUUGGCUGGGGAAAUGGAAAAUGGGGAAUGGAAAGUGGGCGGGGAAAGUGCUC (((((..((((((.(((((.((((((................((((((((....))).)))))........(....).))))....)).))))).).)))))...))))) (-32.89 = -33.00 + 0.11)

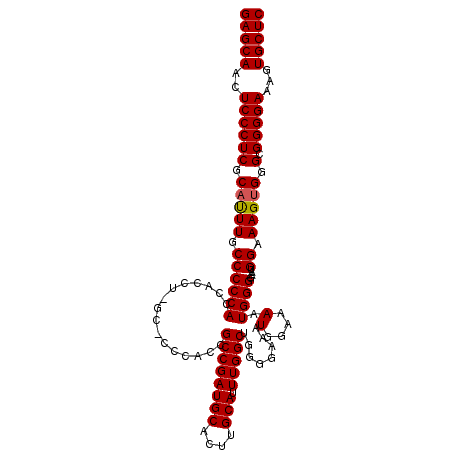

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:30:32 2006