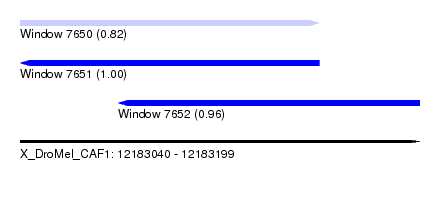
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,183,040 – 12,183,199 |
| Length | 159 |
| Max. P | 0.997327 |

| Location | 12,183,040 – 12,183,159 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.13 |
| Mean single sequence MFE | -19.08 |
| Consensus MFE | -18.15 |
| Energy contribution | -17.96 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.820025 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12183040 119 + 22224390 CCAUUU-UAUCCCUGCCUUCUUUACGUUUUUGUUGUUCAUUUUUUUUCGUAUUUUGGCCAACGCGUAAAAUAUGCUUAAAUAUUUUAAAUUUAAUCACACGCUUCGCUGGCCUCGGCAAA ......-......((((.....((((.....................))))....(((((..(((.......((.((((((.......)))))).)).......))))))))..)))).. ( -20.14) >DroSec_CAF1 9643 115 + 1 CCAUUUUUUUCCCUGCCUUCUUUACGUUUUUGUUGCUCUUU---UUU--UAUUUUGGCCAACGCGUAAAAUAUGCUUAAAUAUUUUAAAUUUAAUCACACGCUUCGCUGGCCUCGGCAAA .............((((......(((....)))........---...--......(((((..(((.......((.((((((.......)))))).)).......))))))))..)))).. ( -18.64) >DroSim_CAF1 12359 118 + 1 CCAUUUUUUUCCCUGCCUUCUUUACGUUUUUGUUGCUCUUUUUUUUU--UAUUUUGGCCAACGCGUAAAAUAUGCUUAAAUAUUUUAAAUUUAAUCACACGCUUCGCUGGCCUCGGCAAA .............((((......(((....)))..............--......(((((..(((.......((.((((((.......)))))).)).......))))))))..)))).. ( -18.64) >DroEre_CAF1 9789 113 + 1 CCAUUU-UUUCCCAGCCUUCUUUGCGUUUUUGUUGUUCUUUUUGU------UUUUGGCCAACGCGUAAAAUAUGCUUAAAUAUUUUAAAUUUAAUCACACGCUUCGCUGGCCUCGGCAAA ......-....(((((.......((((....((((..(.......------....)..))))...((((((((......))))))))...........))))...))))).......... ( -18.90) >consensus CCAUUU_UUUCCCUGCCUUCUUUACGUUUUUGUUGCUCUUUUUUUUU__UAUUUUGGCCAACGCGUAAAAUAUGCUUAAAUAUUUUAAAUUUAAUCACACGCUUCGCUGGCCUCGGCAAA ..............(((......(((....)))......................(((((..(((.......((.((((((.......)))))).)).......))))))))..)))... (-18.15 = -17.96 + -0.19)

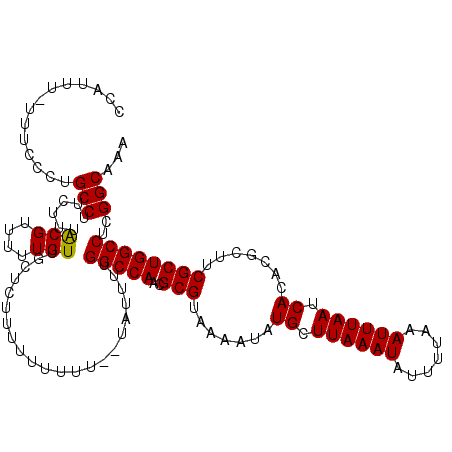
| Location | 12,183,040 – 12,183,159 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.13 |
| Mean single sequence MFE | -27.30 |
| Consensus MFE | -25.98 |
| Energy contribution | -26.23 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.42 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.84 |
| SVM RNA-class probability | 0.997327 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12183040 119 - 22224390 UUUGCCGAGGCCAGCGAAGCGUGUGAUUAAAUUUAAAAUAUUUAAGCAUAUUUUACGCGUUGGCCAAAAUACGAAAAAAAAUGAACAACAAAAACGUAAAGAAGGCAGGGAUA-AAAUGG ((((((..((((((((....(((((.((((((.......)))))).)))))......))))))))....((((.....................)))).....))))))....-...... ( -29.00) >DroSec_CAF1 9643 115 - 1 UUUGCCGAGGCCAGCGAAGCGUGUGAUUAAAUUUAAAAUAUUUAAGCAUAUUUUACGCGUUGGCCAAAAUA--AAA---AAAGAGCAACAAAAACGUAAAGAAGGCAGGGAAAAAAAUGG ((((((..((((((((....(((((.((((((.......)))))).)))))......))))))))......--...---........((......))......))))))........... ( -27.50) >DroSim_CAF1 12359 118 - 1 UUUGCCGAGGCCAGCGAAGCGUGUGAUUAAAUUUAAAAUAUUUAAGCAUAUUUUACGCGUUGGCCAAAAUA--AAAAAAAAAGAGCAACAAAAACGUAAAGAAGGCAGGGAAAAAAAUGG ((((((..((((((((....(((((.((((((.......)))))).)))))......))))))))......--..............((......))......))))))........... ( -27.50) >DroEre_CAF1 9789 113 - 1 UUUGCCGAGGCCAGCGAAGCGUGUGAUUAAAUUUAAAAUAUUUAAGCAUAUUUUACGCGUUGGCCAAAA------ACAAAAAGAACAACAAAAACGCAAAGAAGGCUGGGAAA-AAAUGG (((.(((.((((((((....(((((.((((((.......)))))).)))))......))))))))....------....................((.......))))).)))-...... ( -25.20) >consensus UUUGCCGAGGCCAGCGAAGCGUGUGAUUAAAUUUAAAAUAUUUAAGCAUAUUUUACGCGUUGGCCAAAAUA__AAAAAAAAAGAACAACAAAAACGUAAAGAAGGCAGGGAAA_AAAUGG ((((((..((((((((....(((((.((((((.......)))))).)))))......)))))))).............................(.....)..))))))........... (-25.98 = -26.23 + 0.25)

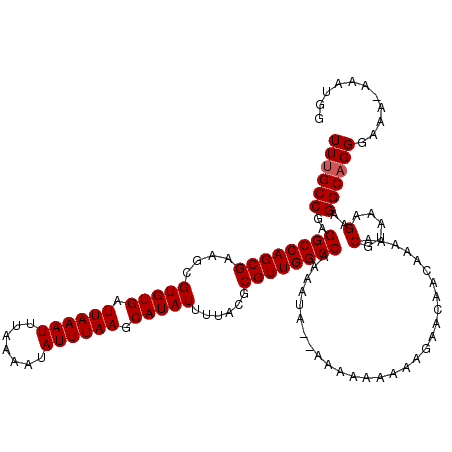

| Location | 12,183,079 – 12,183,199 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.52 |
| Mean single sequence MFE | -29.55 |
| Consensus MFE | -27.10 |
| Energy contribution | -27.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.963787 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12183079 120 - 22224390 AAGCAGCUUCACAUCAUUUGUUUGCAUAUGCAUUGACGUAUUUGCCGAGGCCAGCGAAGCGUGUGAUUAAAUUUAAAAUAUUUAAGCAUAUUUUACGCGUUGGCCAAAAUACGAAAAAAA ..(((((...(((.....)))..))...))).....(((((((.....((((((((....(((((.((((((.......)))))).)))))......)))))))).)))))))....... ( -30.70) >DroSec_CAF1 9683 115 - 1 AAGCAGCUUCACAUCAUUUGUUCGCAUAUGCAUUGACGUAUUUGCCGAGGCCAGCGAAGCGUGUGAUUAAAUUUAAAAUAUUUAAGCAUAUUUUACGCGUUGGCCAAAAUA--AAA---A ................((((((.(((.((((......)))).)))...((((((((....(((((.((((((.......)))))).)))))......))))))))..))))--)).---. ( -28.00) >DroSim_CAF1 12399 118 - 1 AAGCAGCUUCACAUCAUUUGUUUGCAUAUGCAUUGACGUAUUUGCCGAGGCCAGCGAAGCGUGUGAUUAAAUUUAAAAUAUUUAAGCAUAUUUUACGCGUUGGCCAAAAUA--AAAAAAA ................((((((((((.((((......)))).)))...((((((((....(((((.((((((.......)))))).)))))......)))))))).)))))--))..... ( -29.20) >DroEre_CAF1 9828 114 - 1 AAGCAGCUUCACAUCAUUUGUUUGCAUAUGCAUUGACGUAUUUGCCGAGGCCAGCGAAGCGUGUGAUUAAAUUUAAAAUAUUUAAGCAUAUUUUACGCGUUGGCCAAAA------ACAAA ................((((((((((.((((......)))).)))...((((((((....(((((.((((((.......)))))).)))))......))))))))..))------))))) ( -30.30) >consensus AAGCAGCUUCACAUCAUUUGUUUGCAUAUGCAUUGACGUAUUUGCCGAGGCCAGCGAAGCGUGUGAUUAAAUUUAAAAUAUUUAAGCAUAUUUUACGCGUUGGCCAAAAUA__AAAAAAA ..(((((.(((...(((.((....)).)))...))).))...)))...((((((((....(((((.((((((.......)))))).)))))......))))))))............... (-27.10 = -27.10 + -0.00)

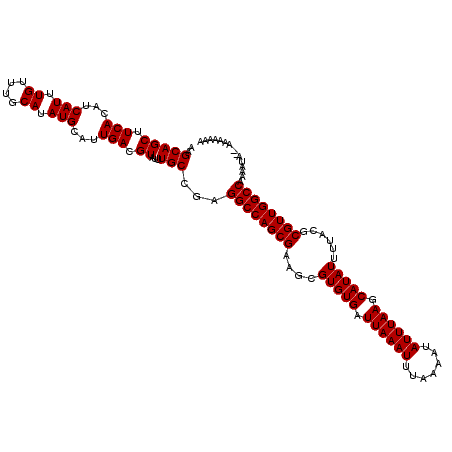
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:30:25 2006