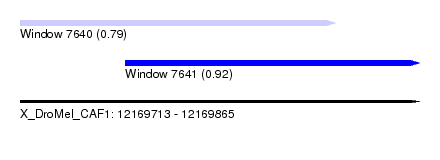
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,169,713 – 12,169,865 |
| Length | 152 |
| Max. P | 0.922744 |

| Location | 12,169,713 – 12,169,833 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.32 |
| Mean single sequence MFE | -16.79 |
| Consensus MFE | -12.50 |
| Energy contribution | -13.17 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.794722 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
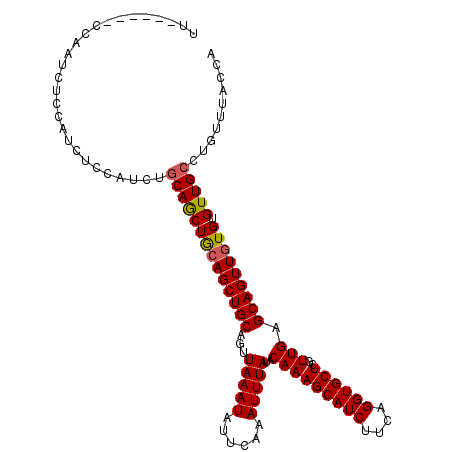
>X_DroMel_CAF1 12169713 120 + 22224390 CAAAGCCAGUGCAUCACCACAUACCCCAUCAUCCCCGCCGUUACCAUCCCAUCCUCUAUCUCCAUCUGCAACUGCAGCUGCAGUUAAAUAUUCAAAUUUAUACAAAGCAUCUUCAGGUGC ....(((.(((......)))..............................................(((((((((....))))))((((......)))).......)))......))).. ( -13.90) >DroEre_CAF1 22052 94 + 1 --------------------CUCAACCAGCAGCUCCUCCAUU------CCAAUCUGCAUCUGCAACUACAGCUACAGCUGCAGUUAAAUAUUCAAAUUUAUACAAAGCAUCUUCAGGUGC --------------------....(((.((((..........------.....))))...(((((((.((((....)))).))))((((......)))).......)))......))).. ( -16.06) >DroYak_CAF1 22707 114 + 1 CAAAGCAAGUGCAUCGCCACCUUCGCCACCAUCACCGCCGUC------GCAAUCUCCAUCUCCAUCUGCAGCUGCAGCUGCAGUUAAAUAUUCAAAUUUAUACAAAGCAUCUUCAGGUGC ....((....)).....(((((..((..........))....------((...............(((((((....))))))).(((((......)))))......))......))))). ( -20.40) >consensus CAAAGC_AGUGCAUC_CCACCUACACCACCAUCACCGCCGUU______CCAAUCUCCAUCUCCAUCUGCAGCUGCAGCUGCAGUUAAAUAUUCAAAUUUAUACAAAGCAUCUUCAGGUGC .................................................................(((((((....))))))).......................(((((....))))) (-12.50 = -13.17 + 0.67)



| Location | 12,169,753 – 12,169,865 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 89.09 |
| Mean single sequence MFE | -25.53 |
| Consensus MFE | -24.01 |
| Energy contribution | -24.23 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922744 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12169753 112 + 22224390 UUACCAUCCCAUCCUCUAUCUCCAUCUGCAACUGCAGCUGCAGUUAAAUAUUCAAAUUUAUACAAAGCAUCUUCAGGUGCUCGUUGAGCAGUUUUGUGUUGCCUGUUUACCA ...........................(((((.((((((((...(((((......)))))..(((((((((....))))))..))).)))))..)))))))).......... ( -22.20) >DroEre_CAF1 22072 106 + 1 UU------CCAAUCUGCAUCUGCAACUACAGCUACAGCUGCAGUUAAAUAUUCAAAUUUAUACAAAGCAUCUUCAGGUGCUCGUUGAGCAGUUGUGUGUUGCCUGUUUACCA ..------.((((..(((.((((((((.((((....)))).)))).................(((((((((....))))))..))).)))).)))..))))........... ( -27.40) >DroYak_CAF1 22747 106 + 1 UC------GCAAUCUCCAUCUCCAUCUGCAGCUGCAGCUGCAGUUAAAUAUUCAAAUUUAUACAAAGCAUCUUCAGGUGCUCGUUGAGCAGUUGUGUGUUGCCUGUUUACCA ..------...................((((((((((((((...(((((......)))))..(((((((((....))))))..))).))))))))).))))).......... ( -27.00) >consensus UU______CCAAUCUCCAUCUCCAUCUGCAGCUGCAGCUGCAGUUAAAUAUUCAAAUUUAUACAAAGCAUCUUCAGGUGCUCGUUGAGCAGUUGUGUGUUGCCUGUUUACCA ...........................((((((((((((((...(((((......)))))..(((((((((....))))))..))).))))))))).))))).......... (-24.01 = -24.23 + 0.23)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:30:14 2006