| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,365,968 – 1,366,076 |
| Length | 108 |
| Max. P | 0.999724 |

| Location | 1,365,968 – 1,366,076 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 73.17 |
| Mean single sequence MFE | -42.25 |
| Consensus MFE | -22.71 |
| Energy contribution | -25.78 |
| Covariance contribution | 3.06 |
| Combinations/Pair | 1.20 |
| Mean z-score | -3.26 |
| Structure conservation index | 0.54 |
| SVM decision value | 3.95 |
| SVM RNA-class probability | 0.999724 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
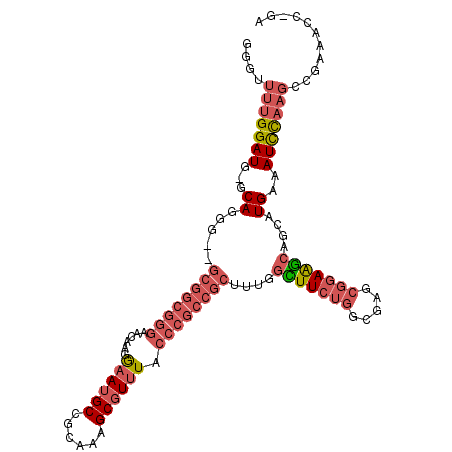
>X_DroMel_CAF1 1365968 108 - 22224390 GGGUUUUGGAUGGGCAGGG--GCGGCGGGUAGAAGGAAUGCCGCAAAGCGUUUACCCGCCGCUUUGGCUUCUGGCGAGCGGAAGCAGGAUGAAAUCCAAGCCGAAACC-GA .((((((((.((((..(((--((((((((((((.((....))((...)).))))))))))))))).(((((((.....))))))).........))))..))))))))-.. ( -52.00) >DroSec_CAF1 26143 108 - 1 GGGUUUUGGAUGGGCAGCG--GCGGCGGGAAGAGGGAAUGCCGCAAAGCGUUUACCCGCCGCUUUGGCUUCUGGCGAGCGGAAGCAGGAUGAAAUCCAAGCCGAAACC-GA .((((((((.(((((((.(--((((((((......((((((......)))))).))))))))))))(((((((.....))))))).........))))..))))))))-.. ( -49.20) >DroEre_CAF1 26600 107 - 1 GGGUUUUGGAUG-GCAGGG--GCGGCGGGAACAGGGAAUGCCACAAAGCGUUUACCCGCCGCUUUGGUUUCUGGCGAGCGGAAGCAGCAUGAAAUCUAAGACGAAACC-GA .(((((((....-...(((--((((((((......((((((......)))))).)))))))))))((((((..((..((....)).))..)))))).....)))))))-.. ( -46.10) >DroAna_CAF1 26085 87 - 1 -------GGAUA-ACAUGGAAACUAUGGAAACAAGAAAAGCCGCAAAGCGUCUUC------------UUUUUGGCG----CAGAUGGCCUGAAAUUCAAGCCGAAAGCUGA -------((...-....(....)..((....)).......))((...(((((...------------.....))))----)...((((.((.....)).))))...))... ( -21.70) >consensus GGGUUUUGGAUG_GCAGGG__GCGGCGGGAACAAGGAAUGCCGCAAAGCGUUUACCCGCCGCUUUGGCUUCUGGCGAGCGGAAGCAGCAUGAAAUCCAAGCCGAAACC_GA ....(((((((...((.....((((((((......((((((......)))))).))))))))....(((((((.....)))))))....))..)))))))........... (-22.71 = -25.78 + 3.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:45:11 2006