
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,154,540 – 12,154,649 |
| Length | 109 |
| Max. P | 0.989053 |

| Location | 12,154,540 – 12,154,649 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 76.09 |
| Mean single sequence MFE | -26.78 |
| Consensus MFE | -16.42 |
| Energy contribution | -16.70 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.61 |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.989053 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12154540 109 + 22224390 AACGGUGACCAAAAAUUAAUUGGAAUUCUCACAUUGGGGAAUUUCAUUGUCUGAUUUUUCAUUGCCCCAGUACACAACAUAUAU-GUACAUACGUAUGUGCGAUUG--UAUG ...((..(..((((((((..((((((((((......)))))))))).....))))))))..)..))...(((((...(((((((-((....)))))))))....))--))). ( -29.10) >DroPse_CAF1 21527 83 + 1 AACGGUGACCAGAAAUUAAUUGGAAUUCUCACAUUGGGGAAUUUCAUUGUCUGAUUUUUCAUGGGCUCUUCAUGUACUAUACA----------------------------- ...((((..((((.......((((((((((......))))))))))...))))......(((((.....))))))))).....----------------------------- ( -17.30) >DroSim_CAF1 9762 101 + 1 AACGGUGACCAAAAAUUAAUUGGAAUUCUCACAUUGGGGAAUUUCAUUGUCUGAUUUUUCAUUGCCCCAGUACACAACAUAU---------ACGUAUGUGUGACUG--UGUG ...((..(..((((((((..((((((((((......)))))))))).....))))))))..)..)).((((.((((.((((.---------...))))))))))))--.... ( -28.40) >DroEre_CAF1 7315 111 + 1 AACGGUGACCAGAAAUUAAUUGGAAUUCUCACAUUUGGGAAUUUCAUUGUCUGAUUUUUCAUUGCCCCAGCACACAACAUACAC-ACACACACACAUGUGUGACUGUGUGUG ...((..(..((((((((..(((((((((((....))))))))))).....))))))))..)..))...........(((((((-(..(((((....)))))..)))))))) ( -35.00) >DroYak_CAF1 7695 110 + 1 AACGGUGACCAGAAAUUAAUUGGAAUUCUCACAUUGGGGAAUUUCAUUGUCUGAUUUUUCAUUGCCCCAGUACACAACAUAC-C-AUACAUGUACAUAUGCAUGUGUGCGUG ...((..(..((((((((..((((((((((......)))))))))).....))))))))..)..)).............(((-(-(((((((((....)))))))))).))) ( -30.80) >DroAna_CAF1 6917 97 + 1 AACGGUGACCAGAAAUUAAUUGGAAUUCUCACAUUGGGGAAUUUCAUUGUCUGAUUUUUCAUGGCCCCCUCCCAGUAUAUACCCCCUAU--AUGUAU-------------UA ...(((.(..((((((((..((((((((((......)))))))))).....))))))))..).))).......((((((((.......)--))))))-------------). ( -20.10) >consensus AACGGUGACCAGAAAUUAAUUGGAAUUCUCACAUUGGGGAAUUUCAUUGUCUGAUUUUUCAUUGCCCCAGCACACAACAUACAC__UAC__ACGUAUGUG_GA_UG__UGUG ...(((((..((((((((..((((((((((......)))))))))).....))))))))..))))).............................................. (-16.42 = -16.70 + 0.28)

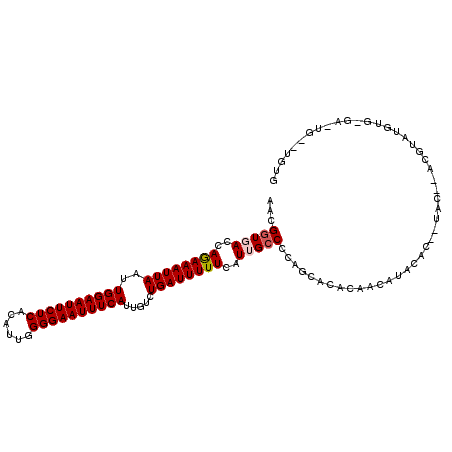
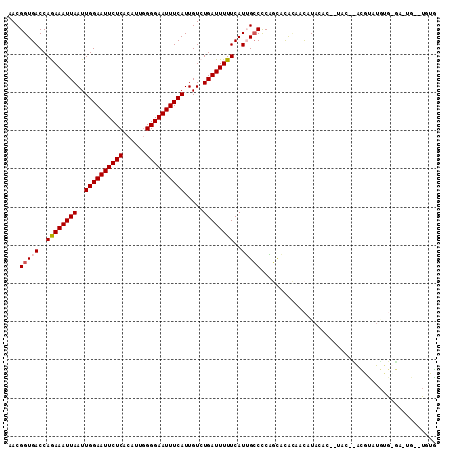
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:30:09 2006