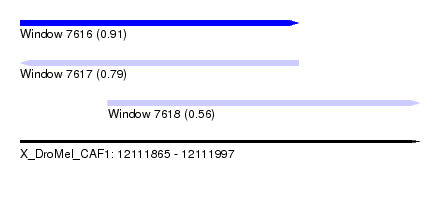
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,111,865 – 12,111,997 |
| Length | 132 |
| Max. P | 0.907942 |

| Location | 12,111,865 – 12,111,957 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.22 |
| Mean single sequence MFE | -26.62 |
| Consensus MFE | -9.85 |
| Energy contribution | -10.80 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.37 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.907942 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12111865 92 + 22224390 -----------GGCAAAAACUUUGCACAUUAAAAUAUGUAAAUUUUAAUUUGAAACAAAAGGGUUGACGGUUGGCA--------GGCAG-UCAG--------CCCUCGCCAACCGUUGAC -----------........((((.((.((((((((......)))))))).)).....)))).((..(((((((((.--------(((..-...)--------))...)))))))))..)) ( -30.60) >DroGri_CAF1 66181 94 + 1 GAAGAGCGCUUCGC-AAAACUUUGCACAUUAAAAUAUGUAAAUUUUAAUUUGAAACAAAAGGGUUGACGUGCUGCU--------GCCAACUCCC--------CCCUUACCG--------- ....(((((..(((-((..((((.((.((((((((......)))))))).)).....))))..))).)).)).)))--------..........--------.........--------- ( -17.00) >DroSec_CAF1 24013 91 + 1 -----------GGCAAAAACUUUGCACAUUAAAAUAUGUAAAUUUUAAUUUGAAACAAAAGGGUUGACGGUUGGCA--------GGCAG-UCAG--------C-CUCGCCAACCGUUGAC -----------........((((.((.((((((((......)))))))).)).....)))).((..((((((((((--------(((..-...)--------)-)).)))))))))..)) ( -33.70) >DroYak_CAF1 36871 92 + 1 -----------GGCAAAAACUUUGCACAUUAAAAUAUGUAAAUUUUAAUUUGAAACAAAAGGGUUGACGGUUGGCA--------GGCAG-UCAG--------CCCUCGCCAAACGUUGAC -----------(((..........((.((((((((......)))))))).)).......(((((((((.(((....--------))).)-))))--------)))).))).......... ( -25.90) >DroAna_CAF1 3422 108 + 1 -----------GGCAGAAAGUUUGCACAUUAAAAUAUGUAAAUUUUAAUUUGAAACAAAAGGGUUGACGGCUGGCAGGACGGCAGGCAG-UCAGGCAGCCCUUCCUCGCCAAAUGUUGAC -----------(((.....((((.((.((((((((......)))))))).))))))..((((((((..(((((.(.........).)))-))...))))))))....))).......... ( -31.80) >DroPer_CAF1 49648 90 + 1 GA--AGCGUUCGGCAAAAACUUUGCACAUUAAAAUAUGUAAAUUUUAAUUUGAAACAAAAGGGUUGACG--UGGCA--------G------------------CCCCGCCAACCGUUGAC ..--((((...(((..........((.((((((((......)))))))).))........((((((...--...))--------)------------------))).)))...))))... ( -20.70) >consensus ___________GGCAAAAACUUUGCACAUUAAAAUAUGUAAAUUUUAAUUUGAAACAAAAGGGUUGACGGUUGGCA________GGCAG_UCAG________CCCUCGCCAACCGUUGAC ...................((((.((.((((((((......)))))))).)).....)))).((..(((..((((................................))))..)))..)) ( -9.85 = -10.80 + 0.95)


| Location | 12,111,865 – 12,111,957 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.22 |
| Mean single sequence MFE | -26.08 |
| Consensus MFE | -9.14 |
| Energy contribution | -10.31 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.35 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.788325 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12111865 92 - 22224390 GUCAACGGUUGGCGAGGG--------CUGA-CUGCC--------UGCCAACCGUCAACCCUUUUGUUUCAAAUUAAAAUUUACAUAUUUUAAUGUGCAAAGUUUUUGCC----------- ((..(((((((((..(((--------(...-..)))--------))))))))))..))..((((((.....((((((((......))))))))..))))))........----------- ( -29.20) >DroGri_CAF1 66181 94 - 1 ---------CGGUAAGGG--------GGGAGUUGGC--------AGCAGCACGUCAACCCUUUUGUUUCAAAUUAAAAUUUACAUAUUUUAAUGUGCAAAGUUUU-GCGAAGCGCUCUUC ---------....(((((--------(((.((((((--------........)))))))))...(((((..((((((((......))))))))..((((....))-))))))).))))). ( -24.60) >DroSec_CAF1 24013 91 - 1 GUCAACGGUUGGCGAG-G--------CUGA-CUGCC--------UGCCAACCGUCAACCCUUUUGUUUCAAAUUAAAAUUUACAUAUUUUAAUGUGCAAAGUUUUUGCC----------- ((..(((((((((.((-(--------(...-..)))--------))))))))))..))..((((((.....((((((((......))))))))..))))))........----------- ( -30.80) >DroYak_CAF1 36871 92 - 1 GUCAACGUUUGGCGAGGG--------CUGA-CUGCC--------UGCCAACCGUCAACCCUUUUGUUUCAAAUUAAAAUUUACAUAUUUUAAUGUGCAAAGUUUUUGCC----------- ((..(((.(((((..(((--------(...-..)))--------)))))).)))..))..((((((.....((((((((......))))))))..))))))........----------- ( -22.60) >DroAna_CAF1 3422 108 - 1 GUCAACAUUUGGCGAGGAAGGGCUGCCUGA-CUGCCUGCCGUCCUGCCAGCCGUCAACCCUUUUGUUUCAAAUUAAAAUUUACAUAUUUUAAUGUGCAAACUUUCUGCC----------- ..........(((.(((((((((((.(.((-(.(....).)))..).))))).........(((((.....((((((((......))))))))..))))))))))))))----------- ( -27.80) >DroPer_CAF1 49648 90 - 1 GUCAACGGUUGGCGGGG------------------C--------UGCCA--CGUCAACCCUUUUGUUUCAAAUUAAAAUUUACAUAUUUUAAUGUGCAAAGUUUUUGCCGAACGCU--UC ......((((((((((.------------------.--------..)).--))))))))..(((((.....((((((((......))))))))..)))))((((.....))))...--.. ( -21.50) >consensus GUCAACGGUUGGCGAGGG________CUGA_CUGCC________UGCCAACCGUCAACCCUUUUGUUUCAAAUUAAAAUUUACAUAUUUUAAUGUGCAAAGUUUUUGCC___________ ((..(((.((((((..............................)))))).)))..))...(((((.....((((((((......))))))))..))))).................... ( -9.14 = -10.31 + 1.17)


| Location | 12,111,894 – 12,111,997 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 85.57 |
| Mean single sequence MFE | -38.18 |
| Consensus MFE | -25.46 |
| Energy contribution | -26.18 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.11 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.559742 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12111894 103 + 22224390 AAUUUUAAUUUGAAACAAAAGGGUUGACGGUUGGCA--------GGCAGUCAG--------CCCUCGCCAACCGUUGACGGCAAUGGCGACGAUGACGACGCCUCCUCAGCCACCUUUA .................(((((((..(((((((((.--------(((.....)--------))...)))))))))..))(((...((((.((....)).))))......))).))))). ( -43.10) >DroSec_CAF1 24042 102 + 1 AAUUUUAAUUUGAAACAAAAGGGUUGACGGUUGGCA--------GGCAGUCAG--------C-CUCGCCAACCGUUGACGGCAAUGGCGACGAUGACGACGCCUCCUCAGCCACCUUAA ..................((((((..((((((((((--------(((.....)--------)-)).)))))))))..))(((...((((.((....)).))))......))).)))).. ( -45.30) >DroEre_CAF1 35564 103 + 1 AAUUUUAAUUUGAAACAAAAGGGUUGACGGUUGGCA--------GGCAGUCAG--------CCCUCGCCAACCGUUGACGGCAAUGGCGACGAUGACGACGCCUCCUCAGCCACCUUAA ..................((((((..(((((((((.--------(((.....)--------))...)))))))))..))(((...((((.((....)).))))......))).)))).. ( -42.20) >DroYak_CAF1 36900 103 + 1 AAUUUUAAUUUGAAACAAAAGGGUUGACGGUUGGCA--------GGCAGUCAG--------CCCUCGCCAAACGUUGACGGCAAUGGCGACGAUGACGACGCUUCCUCAGCCACCUUAA ..................((((((..(((.(((((.--------(((.....)--------))...))))).)))..))(((...((((.((....)).))))......))).)))).. ( -33.70) >DroAna_CAF1 3451 115 + 1 AAUUUUAAUUUGAAACAAAAGGGUUGACGGCUGGCAGGACGGCAGGCAGUCAGGCAGCCCUUCCUCGCCAAAUGUUGACGGCAAUGGCGACGAUGACGACGCC---UCAACCACCUUU- .................(((((((..(((..((((((((.(((..((......)).)))..)))).))))..)))..))((....((((.((....)).))))---....)).)))))- ( -39.50) >DroPer_CAF1 49686 86 + 1 AAUUUUAAUUUGAAACAAAAGGGUUGACG--UGGCA--------G-----------------CCCCGCCAACCGUUGACGGCAAUGGCGACGAUGACGACGCCU---CAACCACCG--- ....................((((..(((--((((.--------.-----------------....))))..)))..))((....((((.((....)).)))).---...)).)).--- ( -25.30) >consensus AAUUUUAAUUUGAAACAAAAGGGUUGACGGUUGGCA________GGCAGUCAG________CCCUCGCCAACCGUUGACGGCAAUGGCGACGAUGACGACGCCUCCUCAGCCACCUUAA ..................((((((..(((.(((((...............................))))).)))..))(((...((((.((....)).))))......))).)))).. (-25.46 = -26.18 + 0.72)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:29:54 2006